
Revisit TBCK-A Pseudo Kinase or a True Kinase
Molecular Biology Molecular BiomarkersCellular BiologyGenomics受け取った 13 Aug 2024 受け入れられた 16 Aug 2024 オンラインで公開された 19 Aug 2024
Focusing on Biology, Medicine and Engineering ISSN: 2995-8067 | Quick Google Scholar
Next Full Text
Revisiting Ice Ages Cycles


受け取った 13 Aug 2024 受け入れられた 16 Aug 2024 オンラインで公開された 19 Aug 2024
Since the initial identification of TBCK (formerly MGC16169) in 2010, significant advances have been made in understanding the role of TBCK mutations in neurodevelopmental disorders such as TBCK encephalopathy. However, the precise function and detailed mechanisms of TBCK remain largely unexplored. Previous studies, including our own, suggest that aberrant expression or mutations in TBCK can impact cell growth, division, and cytoskeleton assembly, contributing to both cancer and neurogenetic diseases. Despite this, the specific domains within TBCK responsible for these functions are still unclear. Notably, mutations in the TBC domain have been implicated in disrupting mTOR pathways, linking TBCK dysfunction to neurogenetic disorders and cancers. Given TBCK’s diverse roles, we have focused on its putative kinase domain. Through comprehensive analysis using tools such as Kinase Tree, AlphaFold2, APE DNA editor, DOG 1., and SMART, we discovered that TBCK lacks the key “D” residue in the conserved “HRD” and “DFG” motifs compared with typical protein kinases PKA and SRC, suggesting TBCK functions as a pseudokinase. Intriguingly, gene ontology analysis from recent RNA-seq data indicates TBCK’s involvement in regulating protein phosphorylation. This suggests that TBCK may influence protein phosphorylation either directly through its potential kinase domain or indirectly via interactions with other proteins. To uncover the full spectrum of TBCK’s roles in neurogenetic disorders and cancer, urgent high-throughput analyses are necessary to identify its interacting partners.
The TBCK gene is a critical player in a range of biological processes. Mutations within the TBCK gene give rise to truncated TBCK protein aggregates, which have been identified as significant contributors to its impaired function, particularly in the context of neurogenetic disorders []. Notably, a specific mutation known as the “Boricua mutation” or p.R126X is strongly associated with a variant of the severe disorder referred to as TBCK-related encephalopathy []. TBCK-related encephalopathy is a rare autosomal recessive neurogenetic condition that presents clinical symptoms such as hypotonia, epilepsy, and intellectual disability, with over 100 documented cases globally []. Recent research has shed light on TBCK’s mutation patterns across different exon loci, providing valuable insights into the genetic basis of this disorder [].
Additionally, the TBCK gene’s implications extend beyond neurogenetic disorders and into cancer progression across various types. Knockdown of TBCK has been shown to elevate levels of phosphorylated Stat3 (pStat3) in A431 cells, suggesting its involvement in the STAT3 signaling pathway []. Furthermore, the long form of TBCK appears to exhibit potential tumor growth-suppressive functions in HeLa cells []. Fusion transcripts, including TBCK-P4HA2 and P4HA2-TBCK, have been reported in a soft tissue angiofibroma case, indicating TBCK’s potential involvement in the development of soft tissue tumors []. In renal cancer cells, miR-1208 targets the 3’UTR of TBCK, reducing TBCK expression and increasing sensitivity to cisplatin and TRAIL treatment []. Moreover, frameshift insertion mutations of TBCK have been detected in 95% of plasma samples from Hepatocellular Carcinoma (HCC) patients, suggesting a potential suppressive role of TBCK in HCC [].
However, the precise mechanisms underlying TBCK’s functions remain insufficiently explored. As previously reported, TBCK comprises protein kinase, TBC, and RHOD domains. The TBC domain is associated with neurogenetic disorders [,,]. Mutations are also identified in the protein kinase domain, underscoring its significance. Notably, TBCK exhibits alternative splicing in various cell lines, primarily in the kinase domain, influencing the length and function of its protein products []. However, our sequence and structural analysis also indicate that TBCK’s kinase domain lacks several critical motifs necessary for kinase activity regulation, which is consistent with previous reports []. Gene ontology analysis from our recent RNA-seq data suggests TBCK’s involvement in regulating protein phosphorylation []. This raises the intriguing possibility that TBCK may participate in protein phosphorylation either directly through its potential kinase domain or indirectly via interactions with binding partners.
TBCK mRNA sequences were downloaded from the NCBI database. Sequence alignment for N terminal sequences of 5 NCBI listed TBCK transcripts using APE DNA editor (https://jorgensen.biology.utah.edu/wayned/ape/) [].
TBCK protein sequence was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/protein/NP_001156907.2?report=fasta) and then input into the online tool SMART (http://smart.embl-heidelberg.de/) [-] to predict the conserved motifs. Next, the Protein Domain Structure Visualization software DOG (Domain Graph, version 1.0) [] was used to visualize all three domains of TBCK in this study.
Detailed motifs and structure for the kinase domain of TBCK were visualized via the online database KinOrtho []. TBCK (336 sequences from different species) and classical Protein kinase sequences (95591 sequences) were aligned and compared. Key motifs for kinase activity were clearly seen in classical protein kinase sequences including GC-rich region, ATP binding β3-Lys, Linker sequence, Catalytic loop, Activation loop, and C/E/F-Helix.
A kinase tree and a dynamic force-directed network for all human kinome and selected categories were displayed via a web application called Coral (http://phanstiel-lab.med.unc.edu/CORAL/) [].
The 3D structures for well-known kinases PKA and SRC were extracted from the public protein database bank (https://www.rcsb.org/) [,]. The 3D structure for TBCK was predicted using AlphaFold2 [-].
In the present study, R “clusterProfiler”, “org.Hs.eg.db”, “enrichplot” and “ggplot2” packages (R version: 3.5.1) were employed to analyze the GO function of the DEGs between sgCtrl and sgTBCK groups [].
Our research group has demonstrated the existence of two alternatively spliced types of TBCK in various cell lines, with splicing events primarily occurring in the kinase domain []. To support this concept, we downloaded five transcript variant sequences for TBCK from the NCBI database and realigned them to illustrate detailed splicing events. For example, variant 2 has a shorter 5’ UTR sequence compared to variant 1 but encodes the same protein product of 893 amino acids (Figure 1). Variant 3 employs an alternate in-frame splice site in the 5’ coding region, skipping a significant portion of exon 6 compared to variant 1, resulting in a shorter protein product (isoform b—854 amino acids) (Figure 2A,B and Figure 3A). The missing protein sequence (161 amino acids to 199 amino acids) in the kinase domain is likely to impact TBCK’s function (Figure 2C). Variant 4, on the other hand, skips two in-frame exons (exon 4 and 5) in the 5’ coding region, generating a shorter protein (isoform c—830 amino acids) (Figure 2A-B and Figure 3B). Similar to isoform b, isoform c lacks a portion of the kinase domain (89 amino acids to 151 amino acids) and is expected to affect TBCK’s function (Figure 2C and 3B). Variant 5 exhibits an alternate exon from intron retention, potentially leading to nonsense-mediated decay (NMD) and translation initiation at a downstream AUG, resulting in a protein with a shorter N-terminus (isoform d—721 amino acids) (Figure 2 and 3C). This isoform also lacks a significant portion of the kinase domain (32 amino acids to 172 amino acids) and is likely to affect TBCK’s function.
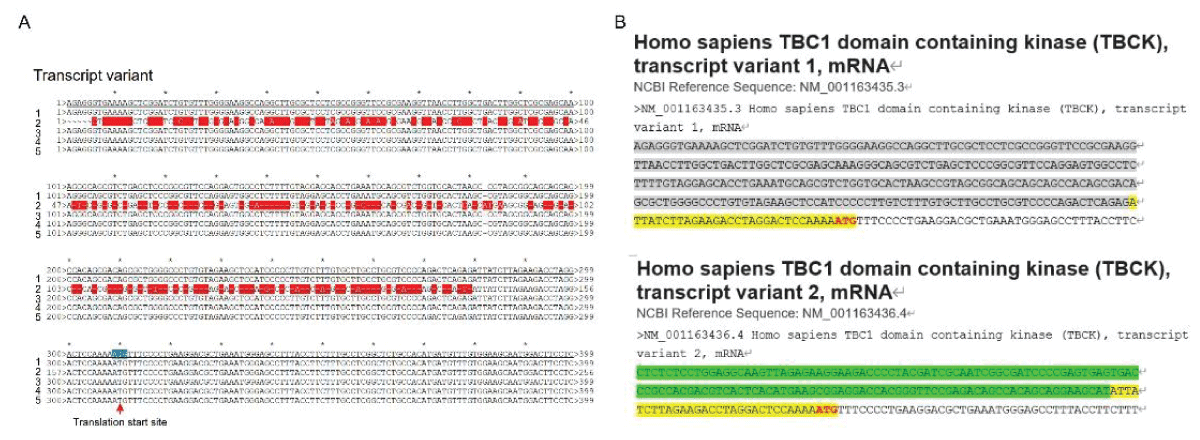 Figure 1: DNA sequence alignment between TBCK transcripts. A. Sequence alignment for N terminal sequences of 5 NCBI listed TBCK transcripts using APE DNA editor (https://jorgensen.biology.utah.edu/wayned/ape/); B. Variant 2 exhibits a shorter 5’ UTR sequence compared to variant 1. But they have the same ORF and encode the same protein product.
Figure 1: DNA sequence alignment between TBCK transcripts. A. Sequence alignment for N terminal sequences of 5 NCBI listed TBCK transcripts using APE DNA editor (https://jorgensen.biology.utah.edu/wayned/ape/); B. Variant 2 exhibits a shorter 5’ UTR sequence compared to variant 1. But they have the same ORF and encode the same protein product.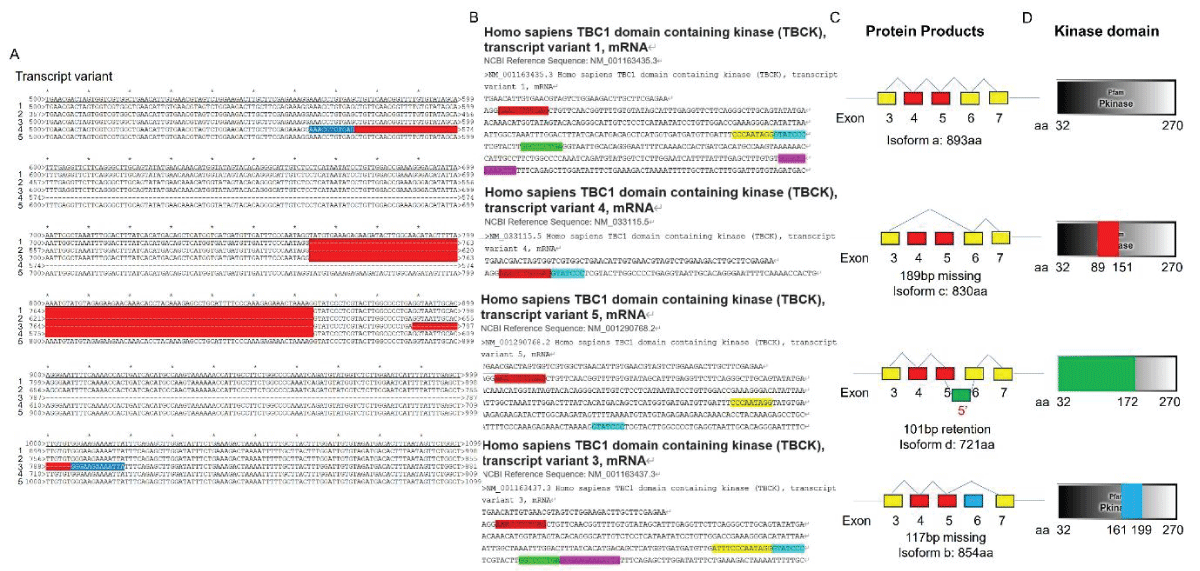 Figure 2: DNA sequence alignment between TBCK transcripts. A. Sequence alignment for N terminal sequences of 5 NCBI listed TBCK transcripts using APE DNA editor (https://jorgensen.biology.utah.edu/wayned/ape/); B. Variant 2 exhibits a shorter 5’ UTR sequence compared to variant 1. But they have the same ORF and encode the same protein product.
Figure 2: DNA sequence alignment between TBCK transcripts. A. Sequence alignment for N terminal sequences of 5 NCBI listed TBCK transcripts using APE DNA editor (https://jorgensen.biology.utah.edu/wayned/ape/); B. Variant 2 exhibits a shorter 5’ UTR sequence compared to variant 1. But they have the same ORF and encode the same protein product.Notably, results from the current analysis using updated database information were consistent with our previous analysis, despite the identification of four more transcripts (f-i) in 2014 []. Nevertheless, the occurrence of splicing events within the kinase domain is apparent. Previous bioinformatic analyses have suggested that TBCK might function as a pseudo kinase, given that its kinase domain lacks several critical motifs for regulating kinase activity, such as GXGXXG, VAIK motifs for ATP binding, and the HRD motif for catalytic activity []. Here, we also did a comprehensive analysis regarding the common features for conserved kinase motifs using two well-known kinases PKA&SRC as references. As shown in Figure S1, a typical kinase comprises 12 subdomains, especially for the key residue “D” in the catalytic loop (Y/HRD) and activation loop (DFG). However, the TBCK putative kinase domain lacked the “D” residue in the catalytic loop (HRA) and activation loop (KFG) (Figure 4A-C). This is consistent with the previous study that TBCK was classified as a pseudo kinase member in the Kinase tree (Figure 3D& Figure 4D) []. Nevertheless, functional assays are required to confirm the extent of its kinase activity and whether it interacts with other protein kinases. Considering our recent data on the involvement of TBCK in the regulation of protein phosphorylation via integrated transcriptome analysis (Figure 3E) [], it is reasonable to believe that TBCK may retain some basal kinase activity or affect other protein kinases. Further investigation is required in this regard (Figure S2).
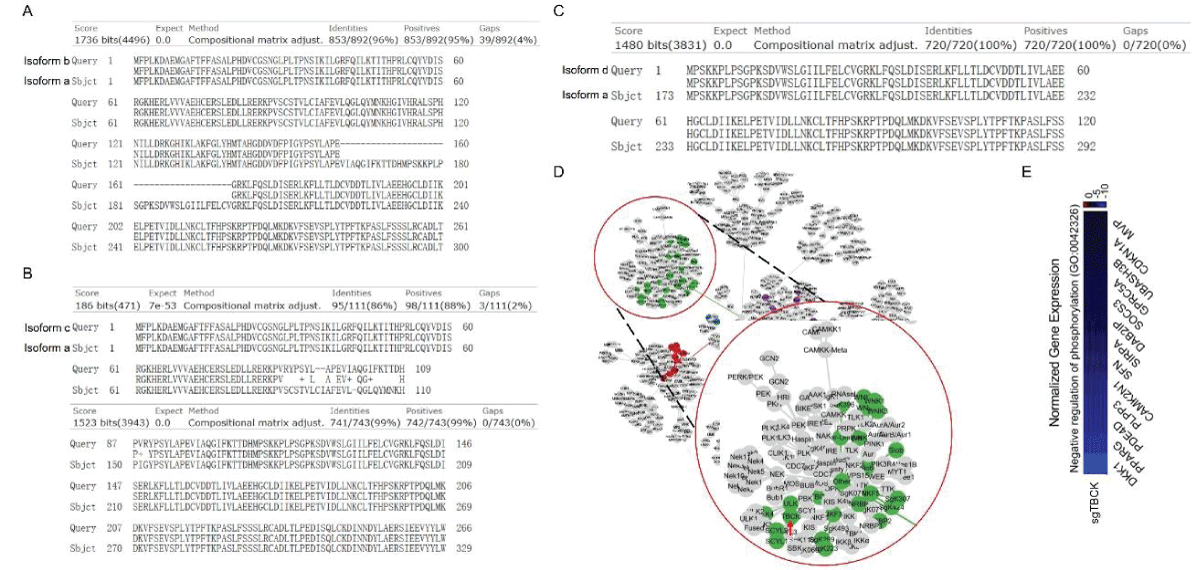 Figure 3: Protein sequence alignment and putative kinase activity of TBCK. A-C. Sequence alignment for 4 TBCK isoforms using online BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&BLAST_SPEC=blast2seq&LINK_LOC=blasttab&LAST_PAGE=blastn&BLAST_INIT=blast2seq); D. a dynamic force-directed network for all human kinome and selected categories were displayed via a web application called Coral; E. Heatmap depicting the differential expression of selected genes in the enriched process: Negative regulation of phosphorylation (GO: 0042326).
Figure 3: Protein sequence alignment and putative kinase activity of TBCK. A-C. Sequence alignment for 4 TBCK isoforms using online BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&BLAST_SPEC=blast2seq&LINK_LOC=blasttab&LAST_PAGE=blastn&BLAST_INIT=blast2seq); D. a dynamic force-directed network for all human kinome and selected categories were displayed via a web application called Coral; E. Heatmap depicting the differential expression of selected genes in the enriched process: Negative regulation of phosphorylation (GO: 0042326).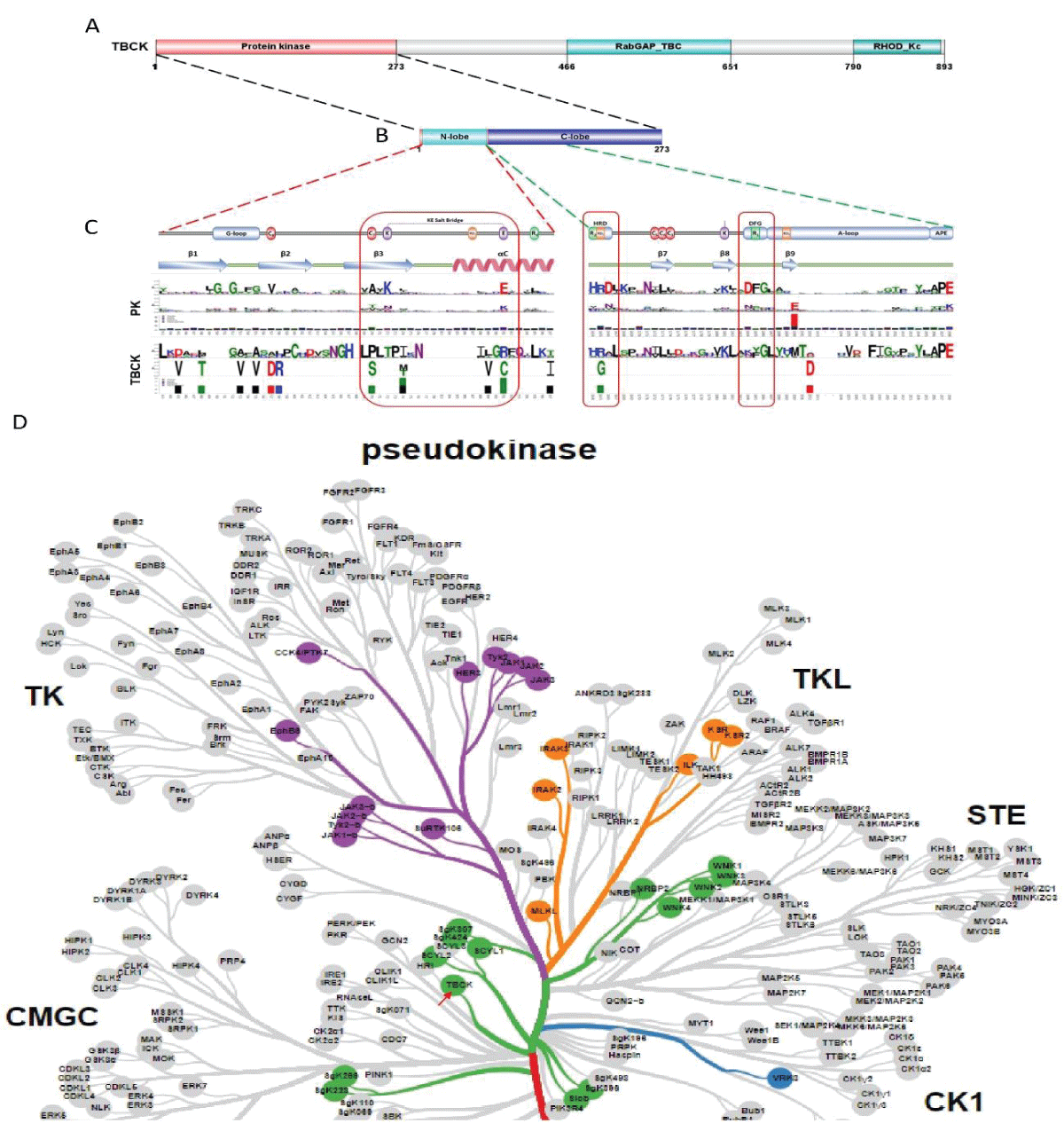 Figure 4: Kinase domain analysis for TBCK. A. A diagram of TBCK including 3 known domains: STYKc; TBC and RHOD; B. N lobe and C-lobe of TBCK kinase domain; C. Detailed motifs and structure for kinase domain of TBCK were visualized via the online database KinOrtho. D. TBCK was listed as a pseudo kinase in a kinase tree.
Figure 4: Kinase domain analysis for TBCK. A. A diagram of TBCK including 3 known domains: STYKc; TBC and RHOD; B. N lobe and C-lobe of TBCK kinase domain; C. Detailed motifs and structure for kinase domain of TBCK were visualized via the online database KinOrtho. D. TBCK was listed as a pseudo kinase in a kinase tree.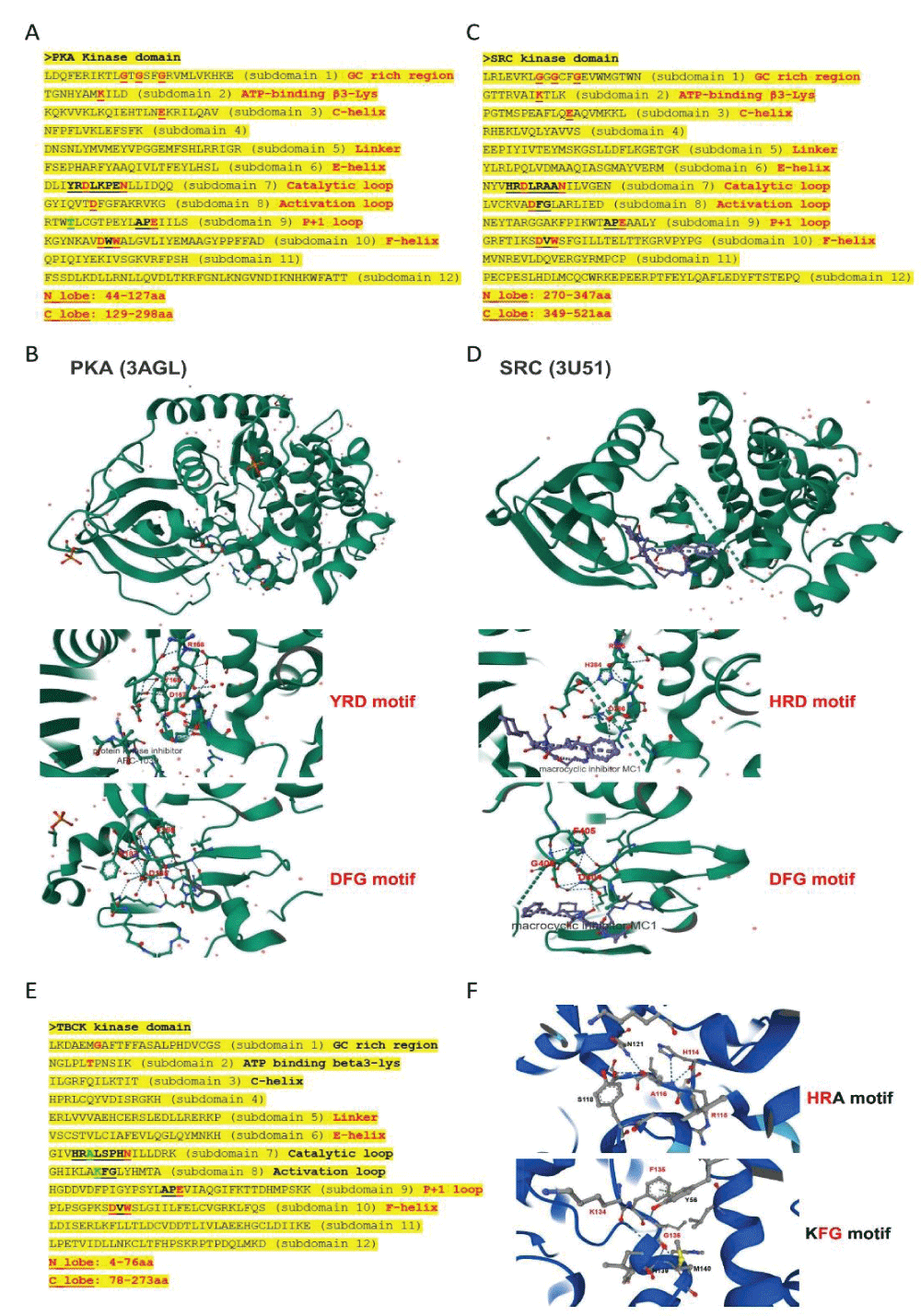 Figure s1: Common structure features for conserved kinase motifs. A. Common sequence features for 12 subdomains of PKA’s kinase domain; B. The 3D structure for PKA as well as the conserved YRD and DFG motifs; C. Common sequence features for 12 subdomains of SRC’s kinase domain; D. The 3D structure for SRC as well as the conserved YRD and DFG motifs; E. The potential Kinase subdomains of TBCK; F. The predicted 3D structures for the HRA and KFG motifs, which missed the key “D” residues.
Figure s1: Common structure features for conserved kinase motifs. A. Common sequence features for 12 subdomains of PKA’s kinase domain; B. The 3D structure for PKA as well as the conserved YRD and DFG motifs; C. Common sequence features for 12 subdomains of SRC’s kinase domain; D. The 3D structure for SRC as well as the conserved YRD and DFG motifs; E. The potential Kinase subdomains of TBCK; F. The predicted 3D structures for the HRA and KFG motifs, which missed the key “D” residues.Our comprehensive analysis of the putative kinase domain of TBCK confirmed that the key residue “D” was missing in the catalytic and activation loops and supported TBCK might be a pseudo kinase. The positive role of TBCK in the regulation of protein phosphorylation suggests that TBCK may retain some basal kinase activity or affect other protein kinases. Urgent high-throughput analyses are essential to screen and identify TBCK’s interacting proteins, which can provide deeper insights into its roles in both neurogenetic disorders and cancer development.
Consent for publication: All authors agree with the manuscript’s content.
Beck-Wödl S, Harzer K, Sturm M, Buchert R, Rieß O, Mennel HD, Latta E, Pagenstecher A, Keber U. Homozygous TBC1 domain-containing kinase (TBCK) mutation causes a novel lysosomal storage disease - a new type of neuronal ceroid lipofuscinosis (CLN15)? Acta Neuropathol Commun. 2018 Dec 27; 6(1):145. doi: 10.1186/s40478-018-0646-6. PMID: 30591081; PMCID: PMC6307319.
Ortiz-González XR, Tintos-Hernández JA, Keller K, Li X, Foley AR, Bharucha-Goebel DX, Kessler SK, Yum SW, Crino PB, He M, Wallace DC, Bönnemann CG. Homozygous boricua TBCK mutation causes neurodegeneration and aberrant autophagy. Ann Neurol. 2018 Jan; 83(1):153-165. doi: 10.1002/ana.25130. PMID: 29283439; PMCID: PMC5876123.
Nair D, Diaz-Rosado A, Varella-Branco E, Ramos I, Black A, Angireddy R, Park J, Murali S, Yoon A, Ciesielski B, O'Brien WT, Passos-Bueno MR, Bhoj E. Heterozygous variants in TBCK cause a mild neurologic syndrome in humans and mice. Am J Med Genet A. 2023 Oct; 191(10):2508-2517. doi: 10.1002/ajmg.a.63320. Epub 2023 Jun 23. PMID: 37353954; PMCID: PMC10524953.
Durham EL, Angireddy R, Black A, Melendez-Perez A, Smith S, Gonzalez EM, Navarro KG, Díaz A, Bhoj EJK, Katsura KA. TBCK syndrome: a rare multi-organ neurodegenerative disease. Trends Mol Med. 2023 Oct; 29(10):783-785. doi: 10.1016/j.molmed.2023.06.009. Epub 2023 Jul 14. PMID: 37455236; PMCID: PMC10868401.
Komurov K, Padron D, Cheng T, Roth M, Rosenblatt KP, White MA. Comprehensive mapping of the human kinome to epidermal growth factor receptor signaling. J Biol Chem. 2010 Jul 2; 285(27):21134-42. doi: 10.1074/jbc.M110.137828. Epub 2010 Apr 26. PMID: 20421302; PMCID: PMC2898331.
Wu J, Li Q, Li Y, Lin J, Yang D, Zhu G, Wang L, He D, Lu G, Zeng C. A long type of TBCK is a novel cytoplasmic and mitotic apparatus-associated protein likely suppressing cell proliferation. J Genet Genomics. 2014 Feb 20; 41(2):69-72. doi: 10.1016/j.jgg.2013.12.006. Epub 2014 Jan 4. PMID: 24576458.
Panagopoulos I, Gorunova L, Viset T, Heim S. Gene fusions AHRR-NCOA2, NCOA2-ETV4, ETV4-AHRR, P4HA2-TBCK, and TBCK-P4HA2 resulting from the translocations t(5;8;17)(p15;q13;q21) and t(4;5)(q24;q31) in a soft tissue angiofibroma. Oncol Rep. 2016 Nov; 36(5):2455-2462. doi: 10.3892/or.2016.5096. Epub 2016 Sep 15. PMID: 27633981; PMCID: PMC5055197.
Kim EA, Jang JH, Sung EG, Song IH, Kim JY, Lee TJ. MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. Int J Mol Sci. 2019 Jul 19; 20(14):3540. doi: 10.3390/ijms20143540. PMID: 31331056; PMCID: PMC6679220.
Gao J, Xi L, Yu R, Xu H, Wu M, Huang H. Differential Mutation Detection Capability Through Capture-Based Targeted Sequencing in Plasma Samples in Hepatocellular Carcinoma. Front Oncol. 2021 Apr 30;11:596789. doi: 10.3389/fonc.2021.596789. PMID: 33996539; PMCID: PMC8120297.
Wu J, Lu G. Multiple functions of TBCK protein in neurodevelopment disorders and tumors. Oncol Lett. 2021 Jan; 21(1):17. doi: 10.3892/ol.2020.12278. Epub 2020 Nov 6. PMID: 33240423; PMCID: PMC7681195.
Chong JX, Caputo V, Phelps IG, Stella L, Worgan L, Dempsey JC, Nguyen A, Leuzzi V, Webster R, Pizzuti A, Marvin CT, Ishak GE, Ardern-Holmes S, Richmond Z; University of Washington Center for Mendelian Genomics; Bamshad MJ, Ortiz-Gonzalez XR, Tartaglia M, Chopra M, Doherty D. Recessive Inactivating Mutations in TBCK, Encoding a Rab GTPase-Activating Protein, Cause Severe Infantile Syndromic Encephalopathy. Am J Hum Genet. 2016 Apr 7; 98(4):772-81. doi: 10.1016/j.ajhg.2016.01.016. Epub 2016 Mar 31. PMID: 27040692; PMCID: PMC4833196.
Davis MW, Jorgensen EM. ApE, A Plasmid Editor: A Freely Available DNA Manipulation and Visualization Program. Front Bioinform. 2022 Feb 4; 2:818619. doi: 10.3389/fbinf.2022.818619. PMID: 36304290; PMCID: PMC9580900.
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004 Jan 1; 32(Database issue):D142-4. doi: 10.1093/nar/gkh088. PMID: 14681379; PMCID: PMC308822.
Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012 Jan; 40(Database issue):D302-5. doi: 10.1093/nar/gkr931. Epub 2011 Nov 3. PMID: 22053084; PMCID: PMC3245027.
Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018 Jan 4; 46(D1):D493-D496. doi: 10.1093/nar/gkx922. PMID: 29040681; PMCID: PMC5753352.
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009 Feb; 19(2):271-3. doi: 10.1038/cr.2009.6. PMID: 19153597.
Huang LC, Taujale R, Gravel N, Venkat A, Yeung W, Byrne DP, Eyers PA, Kannan N. KinOrtho: a method for mapping human kinase orthologs across the tree of life and illuminating understudied kinases. BMC Bioinformatics. 2021 Sep 18; 22(1):446. doi: 10.1186/s12859-021-04358-3. PMID: 34537014; PMCID: PMC8449880.
Metz KS, Deoudes EM, Berginski ME, Jimenez-Ruiz I, Aksoy BA, Hammerbacher J, Gomez SM, Phanstiel DH. Coral: Clear and Customizable Visualization of Human Kinome Data. Cell Syst. 2018 Sep 26;7(3):347-350.e1. doi: 10.1016/j.cels.2018.07.001. Epub 2018 Aug 29. PMID: 30172842; PMCID: PMC6366324.
Pflug A, Rogozina J, Lavogina D, Enkvist E, Uri A, Engh RA, Bossemeyer D. Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions. J Mol Biol. 2010 Oct 15; 403(1):66-77. doi: 10.1016/j.jmb.2010.08.028. Epub 2010 Aug 21. PMID: 20732331.
Georghiou G, Kleiner RE, Pulkoski-Gross M, Liu DR, Seeliger MA. Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles. Nat Chem Biol. 2012 Feb 19;8(4):366-74. doi: 10.1038/nchembio.792. PMID: 22344177; PMCID: PMC3307835.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug; 596(7873):583-589. doi: 10.1038/s41586-021-03819-2. Epub 2021 Jul 15. PMID: 34265844; PMCID: PMC8371605.
Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024 Jan 5; 52(D1):D368-D375. doi: 10.1093/nar/gkad1011. PMID: 37933859; PMCID: PMC10767828.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022 Jan 7; 50(D1):D439-D444. doi: 10.1093/nar/gkab1061. PMID: 34791371; PMCID: PMC8728224.
Wu J, Zhu J, Dynka A, Chang J, Zhang X, Jiang JUN, Lu G, Construction and application of CRISPR-mediated TBCK-Knockout system in multiple human and mouse cell models, (2023).
Liu Y, Yan X, Zhou T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS One. 2013 Aug 19; 8(8):e71349. doi: 10.1371/journal.pone.0071349. PMID: 23977024; PMCID: PMC3747267.
Wu J, Zhu J. Revisit TBCK-A Pseudo Kinase or a True Kinase. August 19, 2024; 2(8): 720-725. IgMin ID: igmin238; DOI: 10.61927/igmin238; Available at: igmin.link/p238
次のリンクを共有した人は、このコンテンツを読むことができます:
1Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, 665 Elm Streets, Buffalo, NY 14203, USA
2Department of Cell Biology and Medical Genetics, School of Basic Medical Science, Shanxi Medical University, Taiyuan 030001, China
Address Correspondence:
Jin Wu, Department of Molecular and Cellular Biology, Roswell Park Comprehensive Cancer Center, 665 Elm Streets, Buffalo, NY 14203, USA, Email: [email protected] | Jianjun Zhu, Department of Cell Biology and Medical Genetics, School of Basic Medical Science, Shanxi Medical University, Taiyuan 030001, China, Email: [email protected]
How to cite this article:
Wu J, Zhu J. Revisit TBCK-A Pseudo Kinase or a True Kinase. August 19, 2024; 2(8): 720-725. IgMin ID: igmin238; DOI: 10.61927/igmin238; Available at: igmin.link/p238
Copyright: © 2024 Wu J, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
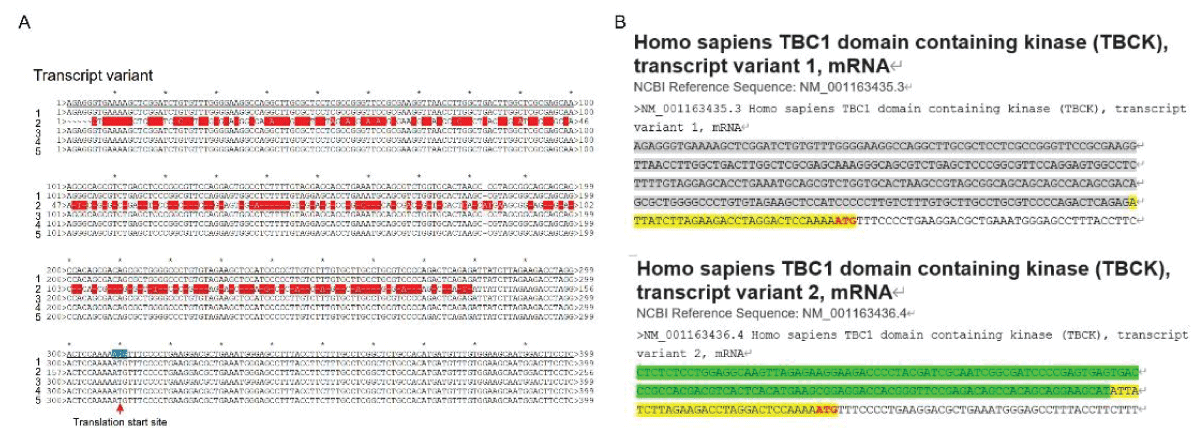 Figure 1: DNA sequence alignment between TBCK transcripts. A...
Figure 1: DNA sequence alignment between TBCK transcripts. A...
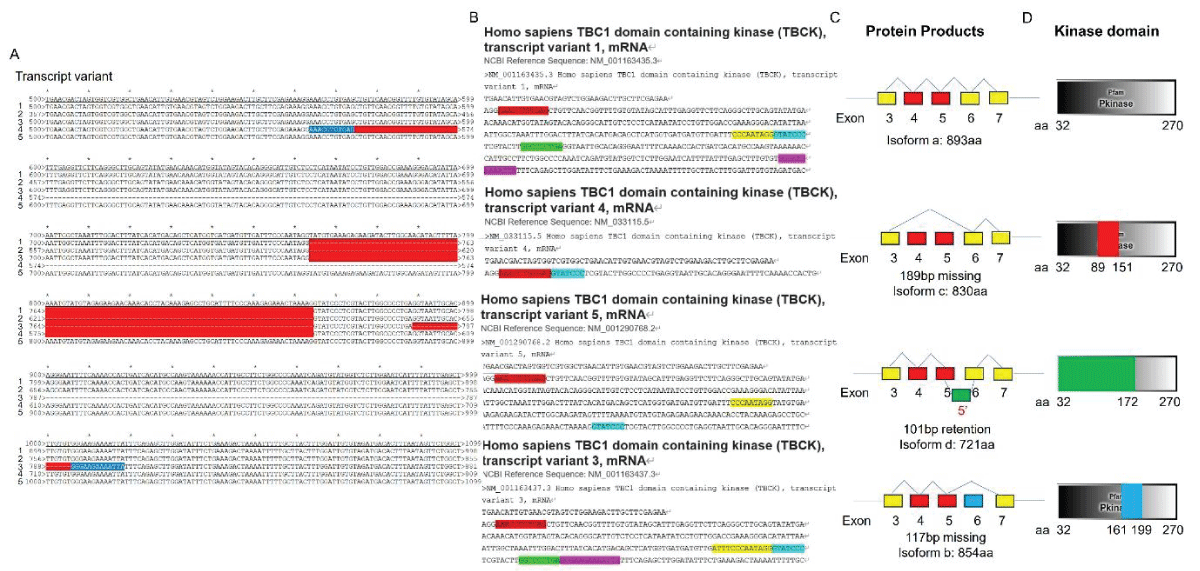 Figure 2: DNA sequence alignment between TBCK transcripts. A...
Figure 2: DNA sequence alignment between TBCK transcripts. A...
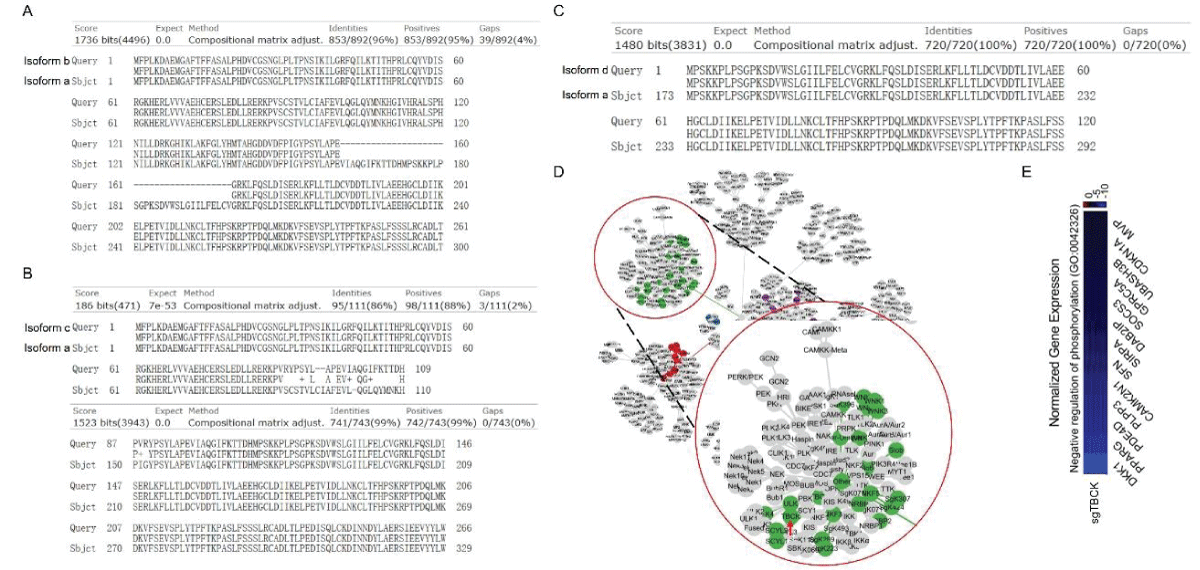 Figure 3: Protein sequence alignment and putative kinase act...
Figure 3: Protein sequence alignment and putative kinase act...
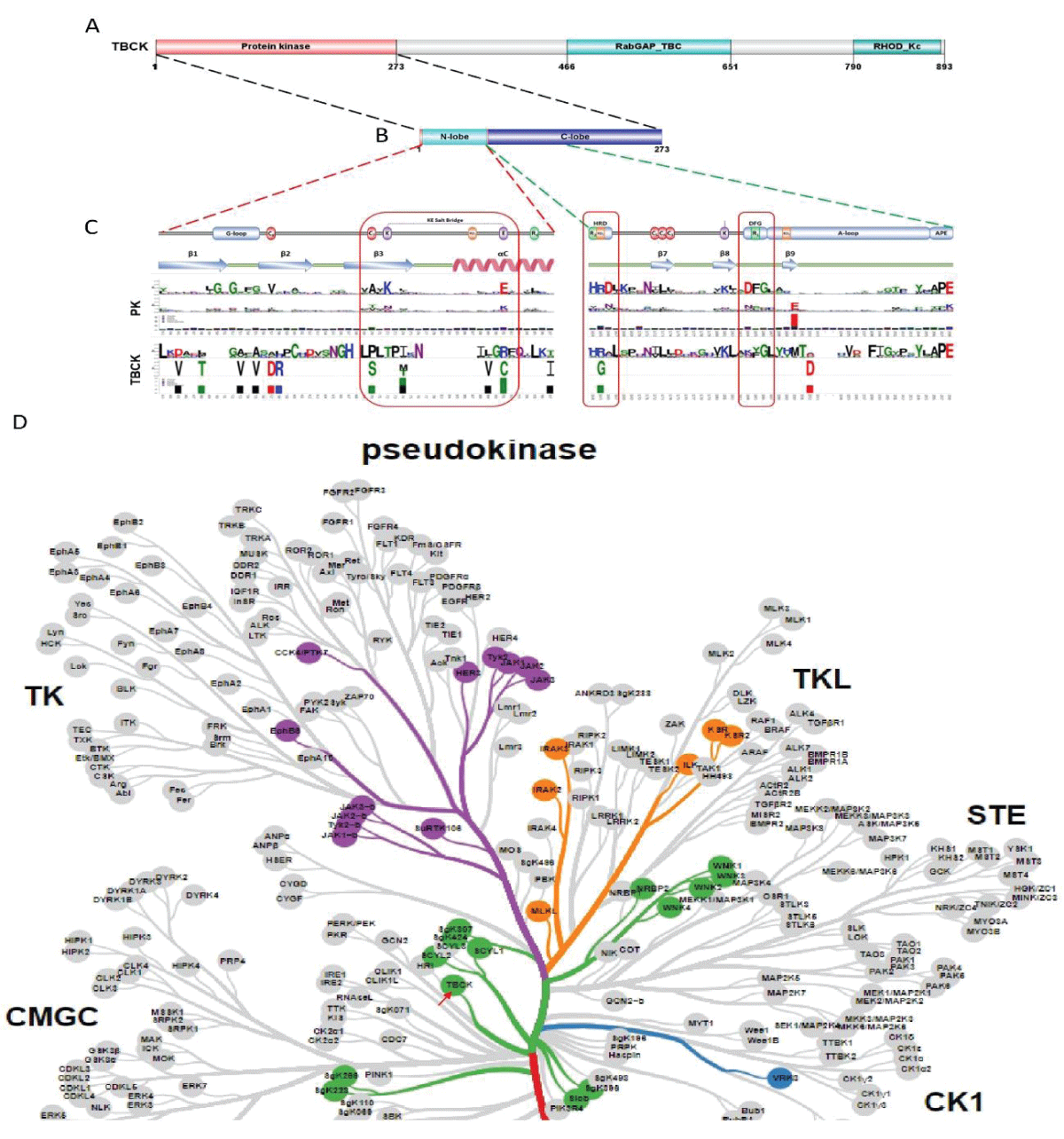 Figure 4: Kinase domain analysis for TBCK. A. A diagram of T...
Figure 4: Kinase domain analysis for TBCK. A. A diagram of T...
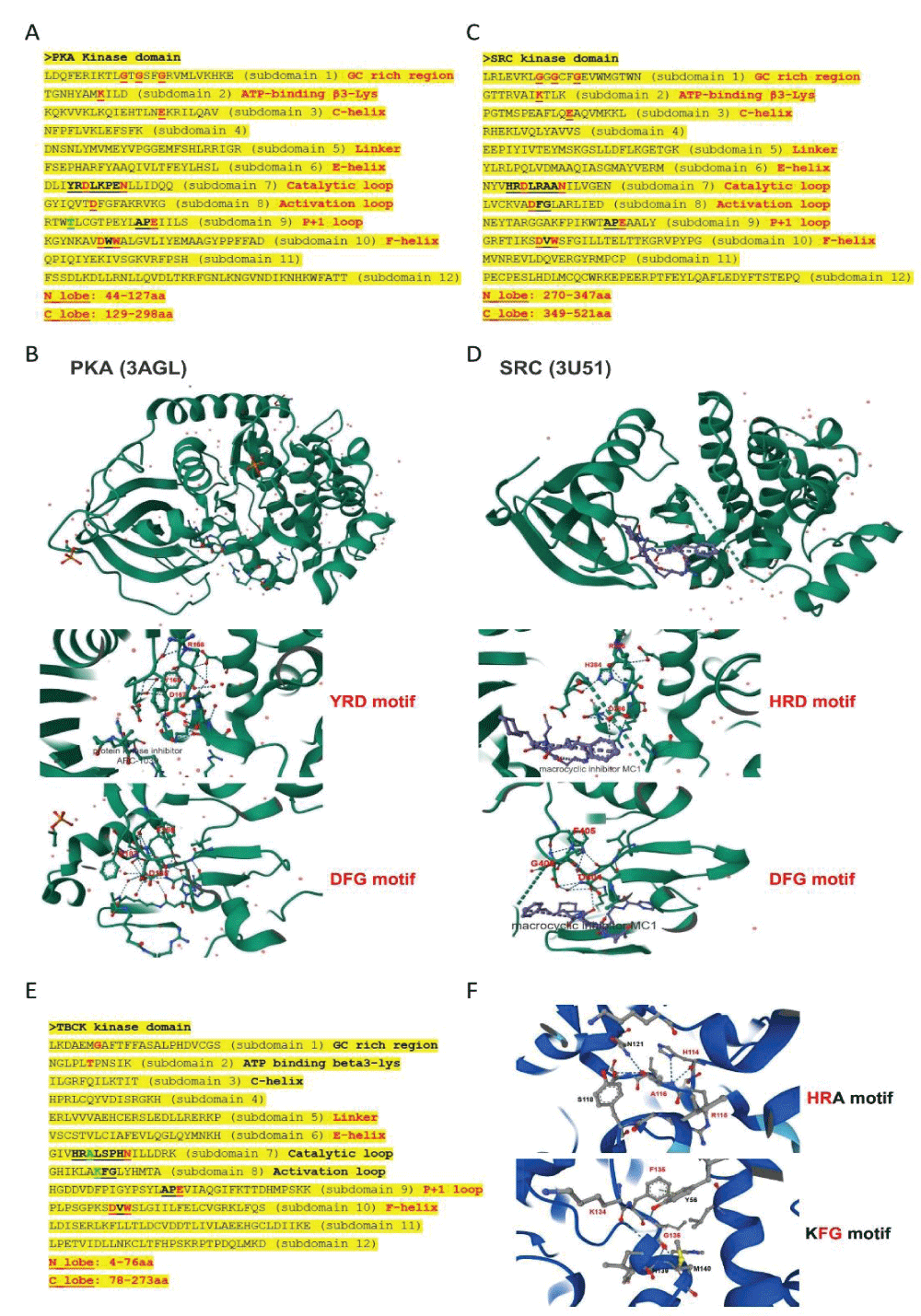 Figure S1: Common structure features for conserved kinase mo...
Figure S1: Common structure features for conserved kinase mo...
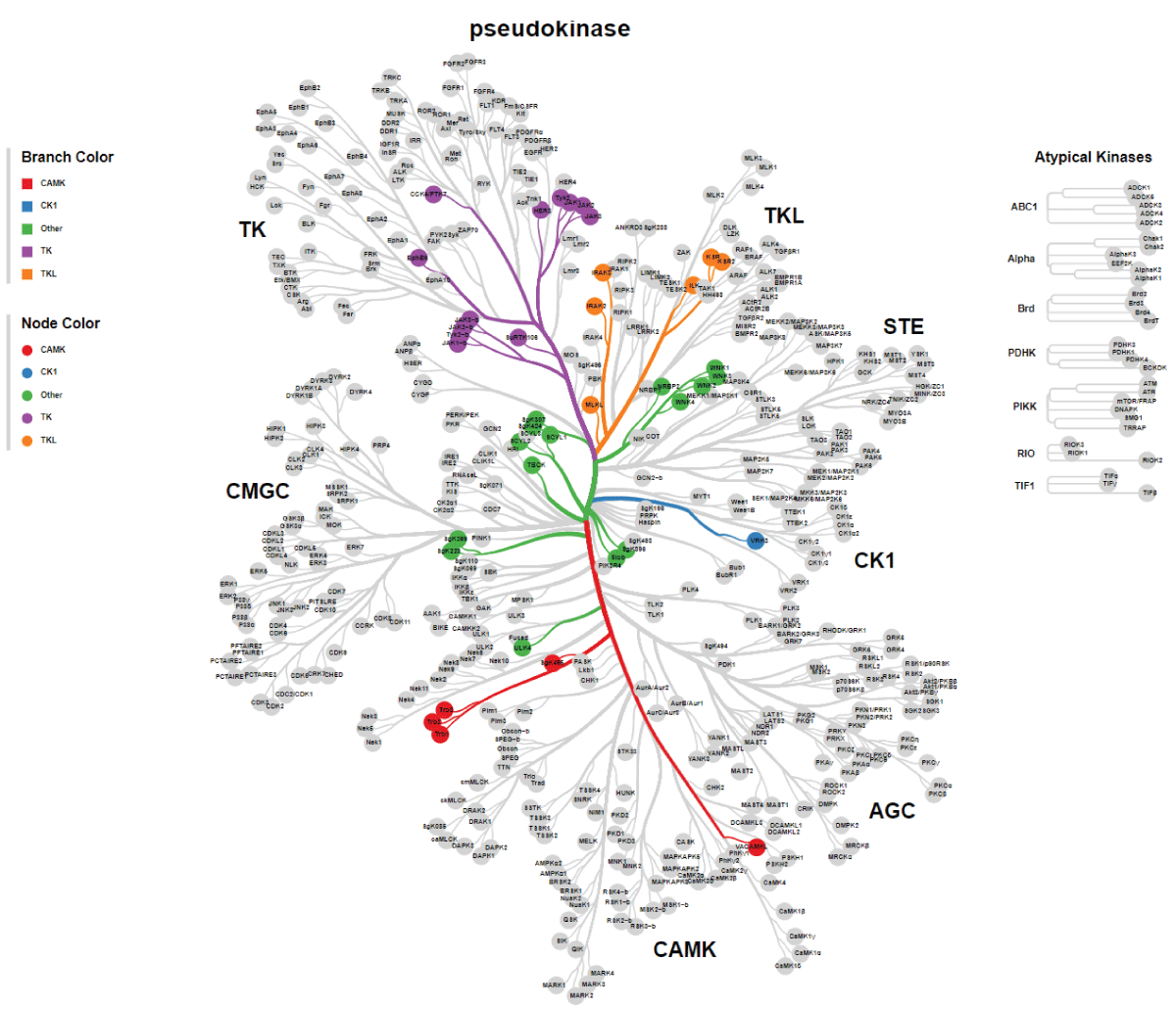 Figure S2: High resolution for the pseudo kinase tree....
Figure S2: High resolution for the pseudo kinase tree....
Beck-Wödl S, Harzer K, Sturm M, Buchert R, Rieß O, Mennel HD, Latta E, Pagenstecher A, Keber U. Homozygous TBC1 domain-containing kinase (TBCK) mutation causes a novel lysosomal storage disease - a new type of neuronal ceroid lipofuscinosis (CLN15)? Acta Neuropathol Commun. 2018 Dec 27; 6(1):145. doi: 10.1186/s40478-018-0646-6. PMID: 30591081; PMCID: PMC6307319.
Ortiz-González XR, Tintos-Hernández JA, Keller K, Li X, Foley AR, Bharucha-Goebel DX, Kessler SK, Yum SW, Crino PB, He M, Wallace DC, Bönnemann CG. Homozygous boricua TBCK mutation causes neurodegeneration and aberrant autophagy. Ann Neurol. 2018 Jan; 83(1):153-165. doi: 10.1002/ana.25130. PMID: 29283439; PMCID: PMC5876123.
Nair D, Diaz-Rosado A, Varella-Branco E, Ramos I, Black A, Angireddy R, Park J, Murali S, Yoon A, Ciesielski B, O'Brien WT, Passos-Bueno MR, Bhoj E. Heterozygous variants in TBCK cause a mild neurologic syndrome in humans and mice. Am J Med Genet A. 2023 Oct; 191(10):2508-2517. doi: 10.1002/ajmg.a.63320. Epub 2023 Jun 23. PMID: 37353954; PMCID: PMC10524953.
Durham EL, Angireddy R, Black A, Melendez-Perez A, Smith S, Gonzalez EM, Navarro KG, Díaz A, Bhoj EJK, Katsura KA. TBCK syndrome: a rare multi-organ neurodegenerative disease. Trends Mol Med. 2023 Oct; 29(10):783-785. doi: 10.1016/j.molmed.2023.06.009. Epub 2023 Jul 14. PMID: 37455236; PMCID: PMC10868401.
Komurov K, Padron D, Cheng T, Roth M, Rosenblatt KP, White MA. Comprehensive mapping of the human kinome to epidermal growth factor receptor signaling. J Biol Chem. 2010 Jul 2; 285(27):21134-42. doi: 10.1074/jbc.M110.137828. Epub 2010 Apr 26. PMID: 20421302; PMCID: PMC2898331.
Wu J, Li Q, Li Y, Lin J, Yang D, Zhu G, Wang L, He D, Lu G, Zeng C. A long type of TBCK is a novel cytoplasmic and mitotic apparatus-associated protein likely suppressing cell proliferation. J Genet Genomics. 2014 Feb 20; 41(2):69-72. doi: 10.1016/j.jgg.2013.12.006. Epub 2014 Jan 4. PMID: 24576458.
Panagopoulos I, Gorunova L, Viset T, Heim S. Gene fusions AHRR-NCOA2, NCOA2-ETV4, ETV4-AHRR, P4HA2-TBCK, and TBCK-P4HA2 resulting from the translocations t(5;8;17)(p15;q13;q21) and t(4;5)(q24;q31) in a soft tissue angiofibroma. Oncol Rep. 2016 Nov; 36(5):2455-2462. doi: 10.3892/or.2016.5096. Epub 2016 Sep 15. PMID: 27633981; PMCID: PMC5055197.
Kim EA, Jang JH, Sung EG, Song IH, Kim JY, Lee TJ. MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. Int J Mol Sci. 2019 Jul 19; 20(14):3540. doi: 10.3390/ijms20143540. PMID: 31331056; PMCID: PMC6679220.
Gao J, Xi L, Yu R, Xu H, Wu M, Huang H. Differential Mutation Detection Capability Through Capture-Based Targeted Sequencing in Plasma Samples in Hepatocellular Carcinoma. Front Oncol. 2021 Apr 30;11:596789. doi: 10.3389/fonc.2021.596789. PMID: 33996539; PMCID: PMC8120297.
Wu J, Lu G. Multiple functions of TBCK protein in neurodevelopment disorders and tumors. Oncol Lett. 2021 Jan; 21(1):17. doi: 10.3892/ol.2020.12278. Epub 2020 Nov 6. PMID: 33240423; PMCID: PMC7681195.
Chong JX, Caputo V, Phelps IG, Stella L, Worgan L, Dempsey JC, Nguyen A, Leuzzi V, Webster R, Pizzuti A, Marvin CT, Ishak GE, Ardern-Holmes S, Richmond Z; University of Washington Center for Mendelian Genomics; Bamshad MJ, Ortiz-Gonzalez XR, Tartaglia M, Chopra M, Doherty D. Recessive Inactivating Mutations in TBCK, Encoding a Rab GTPase-Activating Protein, Cause Severe Infantile Syndromic Encephalopathy. Am J Hum Genet. 2016 Apr 7; 98(4):772-81. doi: 10.1016/j.ajhg.2016.01.016. Epub 2016 Mar 31. PMID: 27040692; PMCID: PMC4833196.
Davis MW, Jorgensen EM. ApE, A Plasmid Editor: A Freely Available DNA Manipulation and Visualization Program. Front Bioinform. 2022 Feb 4; 2:818619. doi: 10.3389/fbinf.2022.818619. PMID: 36304290; PMCID: PMC9580900.
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004 Jan 1; 32(Database issue):D142-4. doi: 10.1093/nar/gkh088. PMID: 14681379; PMCID: PMC308822.
Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012 Jan; 40(Database issue):D302-5. doi: 10.1093/nar/gkr931. Epub 2011 Nov 3. PMID: 22053084; PMCID: PMC3245027.
Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018 Jan 4; 46(D1):D493-D496. doi: 10.1093/nar/gkx922. PMID: 29040681; PMCID: PMC5753352.
Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009 Feb; 19(2):271-3. doi: 10.1038/cr.2009.6. PMID: 19153597.
Huang LC, Taujale R, Gravel N, Venkat A, Yeung W, Byrne DP, Eyers PA, Kannan N. KinOrtho: a method for mapping human kinase orthologs across the tree of life and illuminating understudied kinases. BMC Bioinformatics. 2021 Sep 18; 22(1):446. doi: 10.1186/s12859-021-04358-3. PMID: 34537014; PMCID: PMC8449880.
Metz KS, Deoudes EM, Berginski ME, Jimenez-Ruiz I, Aksoy BA, Hammerbacher J, Gomez SM, Phanstiel DH. Coral: Clear and Customizable Visualization of Human Kinome Data. Cell Syst. 2018 Sep 26;7(3):347-350.e1. doi: 10.1016/j.cels.2018.07.001. Epub 2018 Aug 29. PMID: 30172842; PMCID: PMC6366324.
Pflug A, Rogozina J, Lavogina D, Enkvist E, Uri A, Engh RA, Bossemeyer D. Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions. J Mol Biol. 2010 Oct 15; 403(1):66-77. doi: 10.1016/j.jmb.2010.08.028. Epub 2010 Aug 21. PMID: 20732331.
Georghiou G, Kleiner RE, Pulkoski-Gross M, Liu DR, Seeliger MA. Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles. Nat Chem Biol. 2012 Feb 19;8(4):366-74. doi: 10.1038/nchembio.792. PMID: 22344177; PMCID: PMC3307835.
Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021 Aug; 596(7873):583-589. doi: 10.1038/s41586-021-03819-2. Epub 2021 Jul 15. PMID: 34265844; PMCID: PMC8371605.
Varadi M, Bertoni D, Magana P, Paramval U, Pidruchna I, Radhakrishnan M, et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 2024 Jan 5; 52(D1):D368-D375. doi: 10.1093/nar/gkad1011. PMID: 37933859; PMCID: PMC10767828.
Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022 Jan 7; 50(D1):D439-D444. doi: 10.1093/nar/gkab1061. PMID: 34791371; PMCID: PMC8728224.
Wu J, Zhu J, Dynka A, Chang J, Zhang X, Jiang JUN, Lu G, Construction and application of CRISPR-mediated TBCK-Knockout system in multiple human and mouse cell models, (2023).
Liu Y, Yan X, Zhou T. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS One. 2013 Aug 19; 8(8):e71349. doi: 10.1371/journal.pone.0071349. PMID: 23977024; PMCID: PMC3747267.