
DNA Genetics and UHPLC-Q-TOF-MS Analysis of Phytochemicals for Asparagus racemosus Roots
Molecular Biology Biochemistry受け取った 27 Jun 2024 受け入れられた 08 Jul 2024 オンラインで公開された 10 Jul 2024
Focusing on Biology, Medicine and Engineering ISSN: 2995-8067 | Quick Google Scholar
Previous Full Text
Harnessing Microbes for the Benefit of Man


受け取った 27 Jun 2024 受け入れられた 08 Jul 2024 オンラインで公開された 10 Jul 2024
The medicinal herb Asparagus racemosus has been used in traditional medicine to treat various diseases such as cough, diarrhoea, diabetes, gastric issues, gonorrhoea, headaches, piles, rheumatism, and even lactation enhancement. This study explores the genetic information and phytochemicals of the species native to Gia Lai province, Vietnam before its conservation and cultivation. Five species of A. racemosus were analyzed using the trnL-e/trnL-f regions sequence. A. racemosus roots were extracted by water then Ultra-High-Performance Liquid Chromatography coupled with Quadrupole/Time-Of-Flight Mass Spectrometry (UHPLC-Q-TOF-MS) spectroscopy was used to screen for their phytochemicals. We have confirmed the DNA genetics of A. racemosus species collected in Gia Lai, Vietnam. In water extract of A. racemosus roots, UHPLC-Q-TOF-MS tentatively identified two flavonoids (Quercetin-3-glucuronide, Rutin), five steroidal saponins (Shatavarin I, Shatavarin IV, Shatavarin IX, Asparacoside, Asparanin A), and two steroids (β-sitosterol, Daucosterol). The experimental findings confirm the A. racemosus species for conservation and cultivation in Vietnam and contribute the benefits to the chemical literature of Vietnamese natural flora. A. racemosus should be further studied for pharmaceutical activities.
Asparagus racemosus willd is a well-known medicinal plant belonging to the family Liliaceace []. Generally, A. racemosus plants are found in India, some regions of the Himalayas, and all over the region of Sri Lanka, Asia, Australia, etc. In Vietnam, A. racemosus is found to be distributed in Gia Lai province []. Its tuberous roots are used for bronchitis, constipation, dementia, diabetes, diarrhea, diuretic, dyspepsia, emollient, galactogogue, nervine tonic, rejuvenating, stomachic activities, etc []. The whole plant of A. racemosus including its leaves and roots is especially useful in traditional medicine. As the demand for A. racemosus is constantly on the rise; the supply is inadequate and now considered ‘endangered’ in its natural habitat. It is crucial to carry out its conservation, especially in Vietnam [,].
DNA barcoding is considered an efficient and accurate tool for global species identification []. DNA barcodes that are utilized for the classification of plants mostly belong either to the Internal Transcribed Spacer (ITS)–rDNA region in the nuclear genome, or the rbcL, MatK, psbA–trnH, and atpF–atpH regions in the chloroplast genome [,].
Chemical constituents from the plant roots include several acids such as ferulic acid, isoferulic acid, malic acid, citric acid, asparagusic acid, caffeic acid, and fumaric acid; steroidal glycosides (Shatavarin I-IV), asparaginine, and 9,10-dihydrophenanthrene derivative []. Flowers and mature fruits contain quercetin, rutin, and hyperoside. The phytochemical evaluation of the Asparagus racemosus leaves was performed and showed excessive amounts of steroids and triterpenoids, flavonoids such as diosgenin and quercetin-3-glucuronide [,].
Ultra-High-Performance Liquid Chromatography coupled with Quadrupole/Time-Of-Flight Mass Spectrometry (UHPLC-QTOF-MS/MS) is capable of accurately measuring molecular mass by giving the elemental composition of obtained ions and is widely used in analyzing complex samples due to the high resolution and sensitivity. UHPLC-QTOF-MS was applied to characterize chemical constituents and metabolites in medicinal herbs and obtained considerable results. HPLC-Q-TOF analysis of A. adscendens roots tentatively assigned the possible presence of saponins and spirostanol []. Xue, et al. (2022) have successfully applied the HPLC-Q-TOF-MS/MS to simultaneously quantify five saponin glycosides, asparacoside, shatavarin IX, shatavarin IV, asparanin A and shatavarin V in A. racemosus roots [].
In this research, our first step for the conservation of the plant before its cultivation in the appropriate lands is to confirm the genetics of wild A. racemosus by sequencing psbA-trnH, and ITS1 nuclear rDNA regions. We also study the phytochemistry of A. racemosus roots by UHPLC-Q-TOF-MS spectroscopy.
All reagents were purchased from Sigma-Aldrich Chem. Co. (St. Louis, MO., USA). Deionized water for HPLC; HPLC grade acetonitrile, methanol, and analytical grade formic acid (≥ 98%) were obtained from Fisher (USA).
The roots of A. racemosus from the wild were collected in five different places of Gia Lai Province, Vietnam in October 2023: 1rst set at Kon Lo Khong commune, Kbang district, around point 14005070N, 1080567090E (abbreviation 1); 2nd set at Ia Sao commune, IaGrai district, around point 140048287N, 1070943662E (abbreviation 2); 3rd set at Kon Chieng commune, Mang Yang district, around point 130791426N, 1080276529E (abbreviation 3); 4th set at Yang Nam commune, Krong Chro district, around point 130679035N, 1080512821E (abbreviation 5); and 5th set at IaALe commune, Chu Pu district, around point 13045311N, 107007201E (abbreviation 5). Coordinate was measured by Magellan GPS 315 equipment.
Samples were primarily recognized based on the morphology in the field, then preserved in silica gel-filled bags, and transferred to the Vietnam Academy of Science and Technology for final identification by Dr. Nguyen Sinh Khang, Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology (VAST) before preservation at -30 0C. The voucher specimens have been lodged at the Center for High Technology Development (CHTD, VAST).
All fresh plants were delivered for DNA analysis. All fresh roots were cut off from the plants, dried, powdered, and stored in a fridge for further water extract and HPLC-QTOF chemical analysis.
Total genomic DNA was extracted according to the method of Doyle and Doyle under the local laboratory conditions. Total DNA was extracted from about 100 mg of the plant sample following the CTAB extraction method. The extracted DNA was resuspended in 30 µl miliQ water and a standard 50 ng of the DNA was used for amplification. trnL-e/trnL-f region was amplified using universal primers []. PCR was performed in 25 µL of reaction system containing 7 µL deionized H2O, 12.5 µL of PCR Master mix kit (2×), 1.25 µL of each primer (10 pmol/µL), and 1 µL of DNA template (50 ng). The PCR reaction was performed using PCR Model 9700 (GeneAmp PCR System 9700) for 3 min at 94 °C for denaturation, 35 amplification cycles (45 s at 94 °C for denaturation, 30 s at 55 °C annealing, and 30 s at 72 °C for extension), then 10 min at 72 °C for extension and then held at 4 °C.
PCR products were screened by electrophoresis on 1% agarose gel, and then sequenced at FirstBase Co. Ltd. Raw sequences obtained were assembled and edited by Chromas-Pro 2.1.6 (Technelysium Pty Ltd). All the sequences were then aligned on BLAST, Genbank (http:www.ncbi.nlm.nih.gov/BLAST). Pairwise distance was determined using Mega 7.0 (Kumar, 2016). The phylogenetic trees were constructed using Maximum likelihood and Bayesian inference with a bootstrap value of 1000.
The fresh roots of A. racemosus are washed, sliced, and dried for 5 days in an oven at 40 0C and ground into powder. Dried powder of the roots of A. racemosus was extracted with water (3 times) at room temperature in an ultrasonic 20 kHz and 1 kW extractor. The combined extract was concentrated to dryness under reduced pressure, yielding a residue (brown solid). The residue of the water extract was flushed with nitrogen gas and stored at -20 0C for future use.
Sample analysis by the UHPLC-Q-TOF-MS instrument was investigated with the procedure described previously []. 100.0 (mg) of the extract was accurately weighed into a tube with a cover, and 2.0 (mL) methanol-water (8:2, v/v) solvent was added. The sample was ultrasonicated for 10 min. The sample was filtrated through a 0.45 (μm) filter membrane before injecting for UHPLC-Q-TOF analysis. Sample analysis was performed on an Exion LCTM UHPLC system (AB SCIEX, USA) consisting of an Exion LC degasser, AC pumps, AC autosampler, controller, and AC column oven. Samples were analyzed on a Hypersil GOLD C18 column (150 x 2.1 mm, 3µ) (Thermo Fisher Scientific, USA). The mobile phase, water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B), was run at a flow rate of 0.4 (mL/min) at room temperature. The gradient programming was as follows: 0–4 min, 2% - 20% B; 4-30 min, 20% - 68% B; 30-32 min, 68% - 98% B; 32-40min, 98% B. Sample injection volume was 5.0 (μL).
An AB SCIEX X500R QTOF mass spectrometer (AB SCIEX, USA) with a Turbo V ion source was coupled with the UHPLC system. Mass data were acquired in both negative and positive Electrospray Ionization (ESI) modes. The ESI-MS conditions were set as follows: the ion source temperature, 500 °C; curtain gas, 30 psi; nebulizer gas (GS 1), 45 psi; heater gas (GS 2), 45 psi. For the TOF MS scan, the mass range was set at m/z 70–2000. For the TOF MS/MS scan, the mass range was set at m/z 50–1500. For the negative mode, the ion spray voltage was set at −4.5 kV, the declustering potential (DP) was −70 V, the collision energy (CE) was performed at −20 eV and the Collision Energy Spread (CES) was 10 eV. For the positive mode, the ion spray voltage was set at 5.5 kV, the DP was 80 V, the CE was 20 eV and the CES was 10 eV. All the obtained data were processed by SCIEX OS software version 1.2.0.4122 (AB SCIEX, USA).
For the analysis of plant genetics, at first, its DNA was extracted following the CTAB extraction method. The total genomic DNA of five samples was successfully extracted. Then, the extracted DNA was amplified by PCR technique with appropriate primers. Total DNA extracts were used as the templates to amplify the trnL-e/trnL-f regions (480 bp). The samples gave 100% efficiency for trnL-e/trnL-f - PCR amplification. Electrophoresis of the PCR product from samples showed clear bands with lengths of about 480 bp for trnL-e/trnL-f (Figure 1).
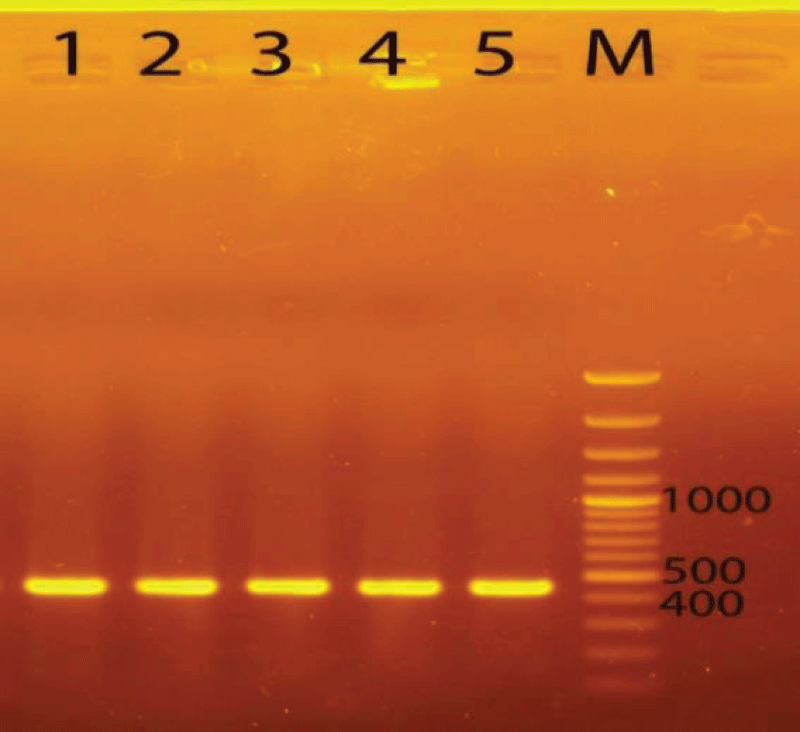 Figure 1: PCR product of DNA regions checked on 1% agarose gel. Abbreviations: 1, 2, 3, 4, 5, M: Marker 100bp plus DNA ladder, trnL-e/trnL-f (GGTTCAAGTCCCTCTATCCC/ATTTGAACTGGTGACACGAG).
Figure 1: PCR product of DNA regions checked on 1% agarose gel. Abbreviations: 1, 2, 3, 4, 5, M: Marker 100bp plus DNA ladder, trnL-e/trnL-f (GGTTCAAGTCCCTCTATCCC/ATTTGAACTGGTGACACGAG).PCR products were directly sequenced and then the obtained sequences were checked for the accuracy of the obtained PCR products by the Blast tool. The sequences were analyzed using Mega 10. The results showed trnL-e/trnL-f regions with 480 bp.
From Table 1 and Figure 2, the molecular phylogenetic tree of five A. racemosus samples and three Asparagus taxa was constructed by maximum likelihood analysis of the conserved regions. The stability of each tree node was tested by bootstrap analysis. Using trnL intron sequence, phylogenetic analysis of genus Asparagus species of Turkey revealed two clades []. In the trnL-e/trnL-f sequence diagram tree, five A. racemosus samples, taxa Asparagus schoberioides JN102165 were classified into two different genealogical branches. The analyzed samples grouped in four branches with similarity between 98,52% - 99,00%, taxa Asparagus racemosus NC047472.1 were closely classified in two different branches. All the referenced taxa are published in GenBank. More phylogenetic data of the Vietnamese A. racemosus species will be exposed when the number of analyzed samples increases. The data will be useful for DNA barcoding based on the authentication of this species.
UHPLC-Q-TOF-MS qualitative analysis
The total ion chromatograms (TICs) of A. racemosus water extract in HPLC-ESI-Q-TOF-MS negative mode are shown in Figure 3.
Some parent and fragment ions were observed in the mass spectra and summarized in Table 2.
Compound (1), at TR = 7.32, in the ESI-QTOF negative mode, yielded a parent ion [M-H]- at m/z 477.1616, and provided fragment ions at m/z 59.0136, m/z 71.0121, m/z 89.0238, m/z 168.0412, and m/z 233.0669. Xue, et al. (2021) found the same compound in the roots of A. racemosus and determined its structure as Quercetin-3-glucuronide []. Thus, compound (1) was tentatively defined as Quercetin-3-glucuronide. Compound (2), at TR = 8.27, in the ESI-QTOF negative mode, yielded a parent ion [M-H]- at m/z 739.2095, and provided major fragment ion at m/z 284.0315. Its structure is determined as Asparanin A []. At TR = 8.47, in the ESI-QTOF negative mode, compound (3) yielded a parent ion at m/z 609.1457 and provided two fragment ions at m/z 112.9856 and m/z 301.0348. Saxena, et al. (2001) found the same compound in A. racemosus and determined its structure as Rutin []. The MS spectra of compound (4) (TR = 9.75 min), yielded a parent ion [M-H]- at m/z 1066.5489 in the negative mode, without any fragmented ion. Compound (4) was determined as Shatavarin I []. Compound 5 (TR = 11.14 min), exhibited parent ion [M-H]- at m/z 901.4792 without any fragmented ion in the ESI-QTOF negative mode. It was assigned to Shatavarin IX [,]. At TR = 12.79, in the ESI-QTOF negative mode, compound (6) yielded a parent ion at m/z 1003.5094 and provided two fragment ions at m/z 112.9856 and m/z 301.0348. Onlom, et al. (2001) found the same compound in A. racemosus and determined its structure as Asparacoside []. Compound (7), at TR = 14.06, in the ESI-QTOF negative mode, yielded a parent ion [M-H]- at m/z 885.4821, without any fragmented ion. Its structure is determined as Shatavarin IV [,]. At TR = 18.93, in the ESI-QTOF negative mode, compound (8) yielded a parent ion at m/z 397.2275 and provided one fragment ion at m/z 96.9595. Ariful (2014) found the same compound in A. racemosus and determined its structure as β-sitosterol []. The MS spectra of compound (9) (TR = 21.08 min), yielded a parent ion [M-H]- at m/z 397.3324 in the negative mode, and provided two fragment ions at m/z 335.3299, m/z 336.3342, and m/z 378.3200. Compound (10) was determined as Daucosterol []. In this study, ultrahigh-resolution liquid chromatography (UPLC) was used to effectively separate the phytoconstituents. A high-resolution mass spectroscopy (Q-TOF-MS/MS) detector was used to determine the parent ion and its fragments which can be compared with databases and literature for the structure of phytoconstituents without the use of their expensive standard compounds. With the use of standard compounds, respective constituents could be quantified by less expensive HPLC-DAD equipment.
In conclusion, from the water extract of A. racemosus roots, we have tentatively identified two flavonoids (Quercetin-3-glucuronide, Rutin), five steroidal saponins (Shatavarin I, Shatavarin IV, Shatavarin IX, Asparacoside, Asparanin A), and two steroids (β-sitosterol, Daucosterol) in the roots of A. racemosus by HPLC-Q-TOF-MS spectroscopy.
This study showed the DNA genetics and trnL-e/trnL-f sequence diagram tree of the Vietnamese plant A. racemosus samples from Gia Lai province. The chemical investigation of their roots was analyzed by UHPLC-Q-TOF-MS spectroscopy with the identification of nine known compounds. More samples collected from different wild forests should be genetically analyzed to show their DNA genetic relationships. And, quantitative analysis of the assigned compounds might be carried out to assess the quality of A. racemosus species. It might be of interest to further investigate the A. racemosus species in isolation of its chemical constituents and pharmaceutical activities.
The project was supported by the grant KHGL-01-22.
Goyal RK, Singh J, Lal H. Asparagus racemosus--an update. Indian J Med Sci. 2003 Sep;57(9):408-14. PMID: 14515032.
Nguyen Van Vu, Nguyen Danh, Tran Minh Đuc. Morphology and molecular structural analysis of Asparagus racemosus wild. From Gia Lai province, Vietnam. Hue Univ J Sci Agric Rural Dev. 2019 Feb;128(3A):83. doi: 10.26459/hueuni-jard.v128i3A.5050.
Guo Y, Liu Z, Wan Y, Zhang Y, Abdu HI, Yang M, et al. Literature analysis on asparagus roots and review of its functional characterizations. Front Nutr. 2023 Apr 17;9:1024190. doi: 10.3389/fnut.2022.1024190. PMID: 37139102; PMCID: PMC10149932.
Bopana N, Saxena S. Asparagus racemosus--ethnopharmacological evaluation and conservation needs. J Ethnopharmacol. 2007 Mar 1;110(1):1-15. doi: 10.1016/j.jep.2007.01.001. Epub 2007 Jan 4. PMID: 17240097.
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2923-8. doi: 10.1073/pnas.0709936105. Epub 2008 Feb 7. PMID: 18258745; PMCID: PMC2268561.
CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12794-7. doi: 10.1073/pnas.0905845106. Epub 2009 Jul 30. PMID: 19666622; PMCID: PMC2722355.
Vassou SL, Nithaniyal S, Raju B, Parani M. Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in Ayurvedic medicine. BMC Complement Altern Med. 2016 Jul 18;16 Suppl 1(Suppl 1):186. doi: 10.1186/s12906-016-1086-0. PMID: 27454470; PMCID: PMC4959393.
Zhang H, Birch J, Pei J, Ma ZF, Bekhit AED. Phytochemical compounds and biological activity in Asparagus roots: a review. Int J Food Sci Technol. 2019; 54:966–77.
Nain S, Singh R, Faujdar S, Paliwal S. Phytochemical Evaluation of Asparagus racemosus and Chlorophytum borivilianum Leaves. Der Pharma Chemica. 2022;14(1).
Battu GS, Ravi Kumar BVV. Phytochemical and antimicrobial activity of leaf extract of Asparagus racemosus Willd. Pharmacognosy Journal. 2010;2:456-463.
Khan KM, Nahar L, Mannan A, Arfan M, Khan GA, Al-Groshi A, et al. Liquid Chromatography Mass Spectrometry Analysis and Cytotoxicity of Asparagus adscendensRoots against Human Cancer Cell Lines. Pharmacogn Mag. 2018 Jan;13(Suppl 4):S890-S894. doi: 10.4103/pm.pm_136_17. Epub 2018 Jan 31. PMID: 29491650; PMCID: PMC5822517.
Xue X, Jin R, Jiao Q, Li X, Li P, Shen G, et al. Differentiation of three Asparagus species by UHPLC-MS/MS based molecular networking identification and chemical profile analysis. J Pharm Biomed Anal. 2022 Sep 20;219:114863. doi: 10.1016/j.jpba.2022.114863. Epub 2022 May 30. PMID: 35785651.
Chen CW, Huang YM, Kuo LY, Nguyen QD, Luu HT, Callado JR, et al. trnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann Bot. 2013 Apr;111(4):663-73. doi: 10.1093/aob/mct004. Epub 2013 Feb 3. PMID: 23380240; PMCID: PMC3605945.
Pham HN, Tran CA, Trinh TD, Nguyen Thi NL, Tran Phan HN, Le VN, et al. UHPLC-Q-TOF-MS/MS Dereplication to Identify Chemical Constituents of Hedera helixLeaves in Vietnam. J Anal Methods Chem. 2022 Aug 8;2022:1167265. doi: 10.1155/2022/1167265. PMID: 35979140; PMCID: PMC9377918.
Altıntaş S, Pakyürek M, Şensoy S, Erez ME, İnal B. Genetic diversity among some asparagus species using rdna its, cpdna trnl intron sequence and screening for antioxidant activity. Pol J Environ Stud. 2019;28(4):2049-2055.
Xue X, Jiao Q, Jin R, Wang X, Li P, Shi S, et al. The combination of UHPLC-HRMS and molecular networking improving discovery efficiency of chemical components in Chinese Classical Formula. Chin Med. 2021 Jul 2;16(1):50. doi: 10.1186/s13020-021-00459-6. PMID: 34215302; PMCID: PMC8254261.
Uddin AV, Anwer B. Asparanin A - Spectroscopic Data of Steroid Glycosides: Stigmastanes, Furostanes, Spirtostanes: Springer US. 2006; 2:1286-1287.
Saxena VK, Chourasia S. A new isoflavone from the roots of Asparagus racemosus. Fitoterapia. 2001 Mar;72(3):307-9. doi: 10.1016/s0367-326x(00)00315-4. PMID: 11295314.
Hayes PY, Jahidin AH, Lehmann R, Penman K, Kitching W, De Voss JJ. Structural revision of shatavarins I and IV, the major components from the roots of Asparagus racemosus. Tetrahedron Lett. 2006 Sep 25;47(39):6965-6969. doi: 10.1016/j.tetlet.2006.07.121.
Onlom C, Nuengchamnong N, Phrompittayarat W, Putalun W, Waranuch N, Ingkaninan K. Quantification of Saponins in Asparagus racemosus by HPLC-Q-TOF-MS/MS. Nat Prod Commun. 2017 Jan;12(1):7-10. PMID: 30549812.
Saran PL, Singh S, Solanki VH, Devi G, Kansara RV, Manivel P. Identification of potential accessions of Asparagus racemosusfor root yield and shatavarin IV content. Heliyon. 2020 Dec 10;6(12):e05674. doi: 10.1016/j.heliyon.2020.e05674. PMID: 33336097; PMCID: PMC7734231.
Hoque A. Phytochemical and Biological Investigation on the Roots of Asparagus rasemosus. 2014. MSc thesis, Bangladesh University of Engineering and Technology
Huong NT, Thuy DN, Trung PV, Hung LN, Van Nam MV. DNA Genetics and UHPLC-Q-TOF‐MS Analysis of Phytochemicals for Asparagus racemosus Roots. IgMin Res. July 10, 2024; 2(7): 558-563. IgMin ID: igmin217; DOI: 10.61927/igmin217; Available at: igmin.link/p217
次のリンクを共有した人は、このコンテンツを読むことができます:
1Center for High Technology Research and Development, Vietnam Academy of Science and Technology, 18-Hoang Quoc Viet, Nghia Do, Cau Giay, Hanoi, Vietnam
2Graduate University of Science and Technology, Vietnam Academy of Science and Technology, 18-Hoang Quoc Viet, Nghia Do, Cau Giay, Hanoi, Vietnam
3Institute of Applied Materials Science, Vietnam Academy of Science and Technology, TL29 Str., Thanh Loc ward, Dist. 12, Ho Chi Minh City, Vietnam
4Environmental and Equipment Technical Corporation, 7/52 Le Trong Tan, Thanh Xuan, Hanoi, Vietnam
Address Correspondence:
Nguyen Thi Huong, Center for High Technology Research and Development, Vietnam Academy of Science and Technology, 18-Hoang Quoc Viet, Nghia Do, Cau Giay, Hanoi, Vietnam, Email: [email protected]
How to cite this article:
Huong NT, Thuy DN, Trung PV, Hung LN, Van Nam MV. DNA Genetics and UHPLC-Q-TOF‐MS Analysis of Phytochemicals for Asparagus racemosus Roots. IgMin Res. July 10, 2024; 2(7): 558-563. IgMin ID: igmin217; DOI: 10.61927/igmin217; Available at: igmin.link/p217
Copyright: © 2024 Huong NT, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
 Figure 1: PCR product of DNA regions checked on 1% agarose g...
Figure 1: PCR product of DNA regions checked on 1% agarose g...
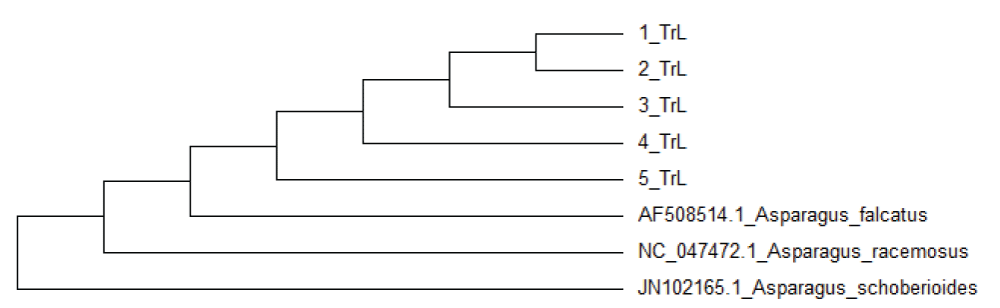 Figure 2: TIC of HPLC-ESI-Q-TOF-MS NEG mode of water extract...
Figure 2: TIC of HPLC-ESI-Q-TOF-MS NEG mode of water extract...
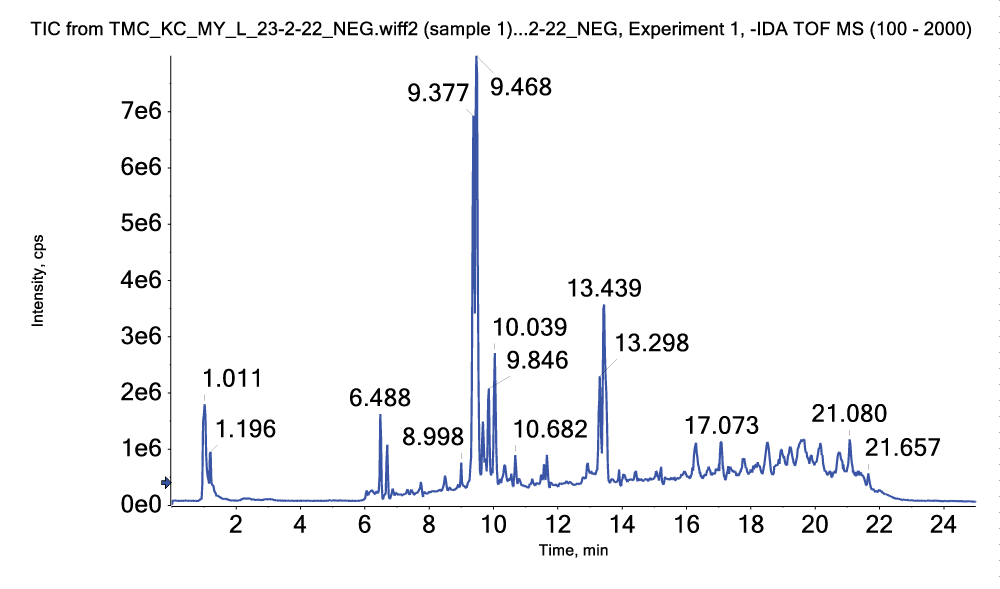 Figure 3: TIC of HPLC-ESI-Q-TOF-MS NEG mode of water extract...
Figure 3: TIC of HPLC-ESI-Q-TOF-MS NEG mode of water extract...
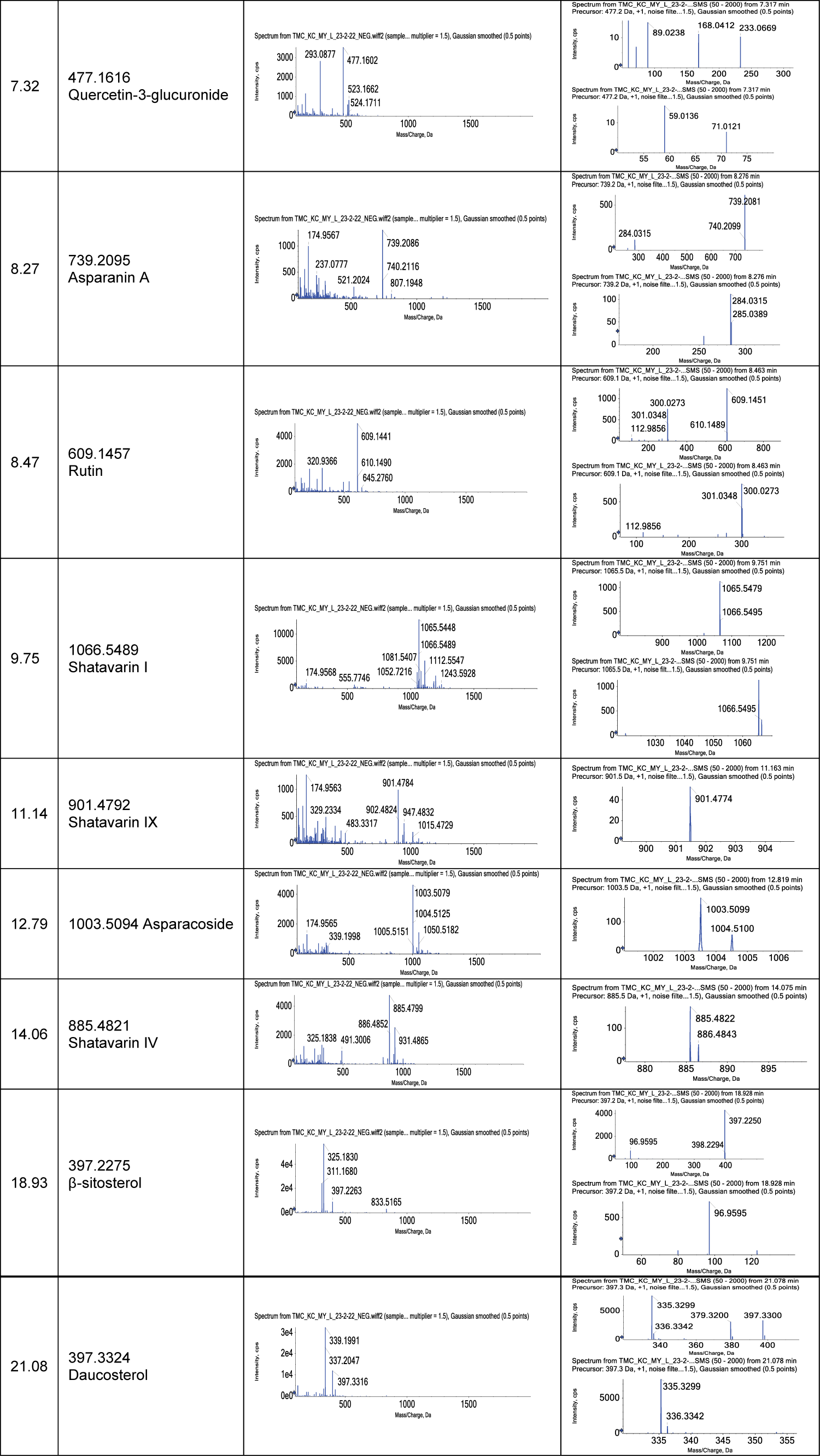 Table 2: HPLC-ESI-Q-TOF NEG assignments of compounds in A. ...
Table 2: HPLC-ESI-Q-TOF NEG assignments of compounds in A. ...
 Table 1: Similarity of the A. racemosus samples....
Table 1: Similarity of the A. racemosus samples....
Goyal RK, Singh J, Lal H. Asparagus racemosus--an update. Indian J Med Sci. 2003 Sep;57(9):408-14. PMID: 14515032.
Nguyen Van Vu, Nguyen Danh, Tran Minh Đuc. Morphology and molecular structural analysis of Asparagus racemosus wild. From Gia Lai province, Vietnam. Hue Univ J Sci Agric Rural Dev. 2019 Feb;128(3A):83. doi: 10.26459/hueuni-jard.v128i3A.5050.
Guo Y, Liu Z, Wan Y, Zhang Y, Abdu HI, Yang M, et al. Literature analysis on asparagus roots and review of its functional characterizations. Front Nutr. 2023 Apr 17;9:1024190. doi: 10.3389/fnut.2022.1024190. PMID: 37139102; PMCID: PMC10149932.
Bopana N, Saxena S. Asparagus racemosus--ethnopharmacological evaluation and conservation needs. J Ethnopharmacol. 2007 Mar 1;110(1):1-15. doi: 10.1016/j.jep.2007.01.001. Epub 2007 Jan 4. PMID: 17240097.
Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2923-8. doi: 10.1073/pnas.0709936105. Epub 2008 Feb 7. PMID: 18258745; PMCID: PMC2268561.
CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009 Aug 4;106(31):12794-7. doi: 10.1073/pnas.0905845106. Epub 2009 Jul 30. PMID: 19666622; PMCID: PMC2722355.
Vassou SL, Nithaniyal S, Raju B, Parani M. Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in Ayurvedic medicine. BMC Complement Altern Med. 2016 Jul 18;16 Suppl 1(Suppl 1):186. doi: 10.1186/s12906-016-1086-0. PMID: 27454470; PMCID: PMC4959393.
Zhang H, Birch J, Pei J, Ma ZF, Bekhit AED. Phytochemical compounds and biological activity in Asparagus roots: a review. Int J Food Sci Technol. 2019; 54:966–77.
Nain S, Singh R, Faujdar S, Paliwal S. Phytochemical Evaluation of Asparagus racemosus and Chlorophytum borivilianum Leaves. Der Pharma Chemica. 2022;14(1).
Battu GS, Ravi Kumar BVV. Phytochemical and antimicrobial activity of leaf extract of Asparagus racemosus Willd. Pharmacognosy Journal. 2010;2:456-463.
Khan KM, Nahar L, Mannan A, Arfan M, Khan GA, Al-Groshi A, et al. Liquid Chromatography Mass Spectrometry Analysis and Cytotoxicity of Asparagus adscendensRoots against Human Cancer Cell Lines. Pharmacogn Mag. 2018 Jan;13(Suppl 4):S890-S894. doi: 10.4103/pm.pm_136_17. Epub 2018 Jan 31. PMID: 29491650; PMCID: PMC5822517.
Xue X, Jin R, Jiao Q, Li X, Li P, Shen G, et al. Differentiation of three Asparagus species by UHPLC-MS/MS based molecular networking identification and chemical profile analysis. J Pharm Biomed Anal. 2022 Sep 20;219:114863. doi: 10.1016/j.jpba.2022.114863. Epub 2022 May 30. PMID: 35785651.
Chen CW, Huang YM, Kuo LY, Nguyen QD, Luu HT, Callado JR, et al. trnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann Bot. 2013 Apr;111(4):663-73. doi: 10.1093/aob/mct004. Epub 2013 Feb 3. PMID: 23380240; PMCID: PMC3605945.
Pham HN, Tran CA, Trinh TD, Nguyen Thi NL, Tran Phan HN, Le VN, et al. UHPLC-Q-TOF-MS/MS Dereplication to Identify Chemical Constituents of Hedera helixLeaves in Vietnam. J Anal Methods Chem. 2022 Aug 8;2022:1167265. doi: 10.1155/2022/1167265. PMID: 35979140; PMCID: PMC9377918.
Altıntaş S, Pakyürek M, Şensoy S, Erez ME, İnal B. Genetic diversity among some asparagus species using rdna its, cpdna trnl intron sequence and screening for antioxidant activity. Pol J Environ Stud. 2019;28(4):2049-2055.
Xue X, Jiao Q, Jin R, Wang X, Li P, Shi S, et al. The combination of UHPLC-HRMS and molecular networking improving discovery efficiency of chemical components in Chinese Classical Formula. Chin Med. 2021 Jul 2;16(1):50. doi: 10.1186/s13020-021-00459-6. PMID: 34215302; PMCID: PMC8254261.
Uddin AV, Anwer B. Asparanin A - Spectroscopic Data of Steroid Glycosides: Stigmastanes, Furostanes, Spirtostanes: Springer US. 2006; 2:1286-1287.
Saxena VK, Chourasia S. A new isoflavone from the roots of Asparagus racemosus. Fitoterapia. 2001 Mar;72(3):307-9. doi: 10.1016/s0367-326x(00)00315-4. PMID: 11295314.
Hayes PY, Jahidin AH, Lehmann R, Penman K, Kitching W, De Voss JJ. Structural revision of shatavarins I and IV, the major components from the roots of Asparagus racemosus. Tetrahedron Lett. 2006 Sep 25;47(39):6965-6969. doi: 10.1016/j.tetlet.2006.07.121.
Onlom C, Nuengchamnong N, Phrompittayarat W, Putalun W, Waranuch N, Ingkaninan K. Quantification of Saponins in Asparagus racemosus by HPLC-Q-TOF-MS/MS. Nat Prod Commun. 2017 Jan;12(1):7-10. PMID: 30549812.
Saran PL, Singh S, Solanki VH, Devi G, Kansara RV, Manivel P. Identification of potential accessions of Asparagus racemosusfor root yield and shatavarin IV content. Heliyon. 2020 Dec 10;6(12):e05674. doi: 10.1016/j.heliyon.2020.e05674. PMID: 33336097; PMCID: PMC7734231.
Hoque A. Phytochemical and Biological Investigation on the Roots of Asparagus rasemosus. 2014. MSc thesis, Bangladesh University of Engineering and Technology