
A Machine Learning-based Method for COVID-19 and Pneumonia Detection
Machine Learning Biomedical Engineering受け取った 05 Jun 2024 受け入れられた 04 Jul 2024 オンラインで公開された 05 Jul 2024
ISSN: 2995-8067 | Quick Google Scholar
Next Full Text
From Motor Augmentation to Body Enhancement, the Harmony between Human Body and Artificial Parts


受け取った 05 Jun 2024 受け入れられた 04 Jul 2024 オンラインで公開された 05 Jul 2024
Pneumonia is described as an acute infection of lung tissue produced by one or more bacteria, and Coronavirus Disease (COVID-19) is a deadly virus that affects the lungs of the human body. The symptoms of COVID-19 disease are closely related to pneumonia. In this work, we identify the patients of pneumonia and coronavirus from chest X-ray images. We used a convolutional neural network for spatial feature learning from X-ray images. We experimented with pneumonia and coronavirus X-ray images in the Kaggle dataset. Pneumonia and corona patients are classified using a feed-forward neural network and hybrid models (CNN+SVM, CNN+RF, and CNN+Xgboost). The experimental findings on the Pneumonia dataset demonstrate that CNN detects Pneumonia patients with 99.47% recall. The overall experiments on COVID-19 x-ray images show that CNN detected the COVID-19 and pneumonia with 95.45% accuracy.
Coronavirus disease (COVID-19) is a deadly virus that the World Health Organization confirmed an epidemic on 11 March 2020 due to its global spread []. The Coronavirus is extremely dangerous because it can reproduce for up to two weeks without causing illness []. COVID-19 symptoms include a high fever, dry cough, stoutness of breath, tiredness, sore throat, aches and pains, and only a few people will observe runny nose, nausea, and diarrhea []. Pneumonia is an acute infection of lung tissue produced by one or more bacteria. The symptoms of COVID-19 disease are closely related to pneumonia []. Radiologists may struggle to detect pneumonia in chest radiography []. In X-ray images, the appearance of pneumonia is usually unclear, can overlap with another diagnosis, and can mimic a variety of other minor anomalies []. These inconsistencies result in significant variation in the diagnosis of pneumonia among radiologists []. Pneumonia and COVID-19 can be identified using various methods, including chest X-rays and CT scans []. Many current studies [,] on using chest X-rays to diagnose COVID-19 have shown significant results. Despite being based on small datasets, the findings suggest high sensitivity to COVID-19. The COVID-19 group’s disease stage is unknown; all we know is that chest X-rays are abnormal []. This is one of our data’s limitations. To classify COVID-19 from the X-ray dataset, Hemdan, et al. suggested seven Convolutional Neural Network models containing modified Google MobileNet and VGG19 []. Narin, et al. [] offered a hybrid method that used InceptionV3, ResNet50, and ResNetV2 networks to distinguish between normal chest pictures and those with COVID-19 diagnostic images. Examines in [] people who were brought to a hospital in Wuhan, China, after testing positive for COVID-19 pneumonia. They separated CT scan patients into groups and then analyzed and compared the image features and distribution to detect COVID-19 diseases. Our work used CNN and hybrid models to classify pneumonia and corona patients. First feature learning is performed using CNN, and the output of the first dense layer is passed to the SVM, RF, and xgboost. The classification of pneumonia is performed using this hybrid approach. In COVID-19 detection, we apply PCA to CT scan images and then pass these components to the SVM for classification.
The COVID-19 virus now affects all humans on Earth. COVID-19 disease has symptoms that are related to pneumonia. The primary purpose of this study is to distinguish COVID-19 patients from pneumonia patients. As a result, new textural features and a hybrid model of (CNN+SVM, CNN+RF, CNN+XGBOOST) chest image classification method were presented.
The presented work is unique in two aspects. One component is the method for optimizing CNN hyperparameters; the other uses CNN with RF, SVM, and XGBOOST. We also compare the performance of CNN feature learning with PCA feature decomposition in COVID-19 detection. The main work of this paper is listed below: 1): Performance Comparison of different optimizers in training of CNN for classification of pneumonia and COVID-19. 2): Performance Comparison of CNN by using different traditional machine learning methods with a feed-forward neural network. 3): Performance comparison of CNN feature learning with PCA feature decomposition method.
The paper is structured as follows: Section 2 mentions related works. Section 3 specifies the designed methodology. Section 4 analyzes the results. Section 5 states the conclusion and future work.
To ease patient suffering, doctors must be able to provide accurate diagnoses and identify the source of disease problems promptly. Image processing and deep learning algorithms have produced promising results in diagnosing and processing biomedical images. This part provides an overview of several major contributions from the current literature.
Pneumonia is one of the diseases that has become more common. Togacar, et al. [] examined lung X-ray images. To complete a specific task, use CNN for feature extraction and models such as AlexNet, VGG-16, and VGG-19 as convolutional neural network models. By decreasing the number of deep features in each deep model, the total number of deep features is decreased. The classification process was then done using decision trees, KNN, SVM, and LDA. According to findings, deep features gave consistent features for identifying pneumonia. Liang, et al. [] suggested a novel deep-learning approach for classifying child pneumonia images that combined partial and dilated convolution. The proposed method used residual structure and dilated convolution to tackle the model’s overfitting and degradation issues. The study in [] offers a deep learning-based technique for detecting and classifying Chest X-Ray pneumonia images. A mask-RCNN detection model, which contains global and local pixel segmentation features, is used. The model’s usefulness and reliability are validated by its good performance on the chest X-ray images dataset.
Bandyopadhyay, et al. [] stated a new model that used LSTM-GRU to classify COVID-19 cases as confirmed, released, negative, or dead. The Presented approach achieved an accuracy rate of 87% for verified cases, 67.8% for negative cases, 62% for dead patients, and 40.5% for released patients. Wang, et al. [] presented COVID-Net, CNN developed from chest X-ray images for COVID-19 detecting patients. Images in the dataset are labeled as COVID-19, V_pneumonia, B_pneumonia, and N_ormal. COVID Net obtained an accuracy of 83.5%, whereas non-COVID patients accuracy achieved 92.4%. Kumar, et al. [] described a technique for detecting COVID-19 via X-ray images based on a deep Convolutional network. The model is trained using data from GitHub; Kaggle obtained an accuracy of 95.38% in recognizing COVID-19 patients. Oztrk, et al. [] introduced DarkCovidNet, CNN based on the DarkNet model for identification of COVID-19 using chest X-ray dataset. Multi-class diagnoses are proposed in DarkCovid for multi-class; classification accuracy is 87.02 %, and for two classes, 98.08%. Ioanis, et al. [] used two datasets to train various pre-trained models. One is a group of 1,427 X-ray images, 504 normal images, 700 pneumonia cases, and 224 COVID-19_cases. The second dataset contains images of 504 normal cases, 714 confirmed viral pneumonia, and 224 Confirm_COVID-19 patients. The model gives an accuracy for two classes of 98.75%, and for three classes, accuracy is 93.48%.
In our work, we select the dataset from Kaggle and perform preprocessing. Then, we use CNN and a hybrid method for COVID-19 and pneumonia classification. In the fourth step, we evaluate the performance of our proposed method using a different evaluation matrix. Figure 1 shows the workflow of our proposed work.
In this study, we will use two datasets. One dataset is taken from the Kaggle Competition []. It contains 5863 images of chest X-rays and is divided into three sections: training, validation, and testing.
Figure 2 shows the normal and Pneumonia Xray images from the Kaggle dataset.
This study performed the grayscale conversion as radiographic images are typically more informative in this format. It used Gaussian blur, median blur, or bilateral filtering to reduce noise in the photos. Pixel normalization is also performed. To ensure consistency, to a standard range (e.g., 0-255). We performed data augmentation using API’s ImageDataGenerator function, which is performed using Keras. We also scaled the images using the Python reshape function.
We proposed a Hybrid approach for disease classification, which consists of two models, CNN+SVM, CNN+RF, and CNN+XGBOOST model, to detect pneumonia from images of chest X-rays. In dataset 1, we have a binary class problem in which two classes are pre-defined. One is Normal images class and the other is Pneumonia Images. We will experiment with two datasets with the help of our hybrid approach and evaluate the results of both datasets. This work will use another optimizer like RMSprop stochastic gradient descent with momentum. We will perform a comparison analysis of different optimizers on these two datasets. This study used the K fold cross-validation to validate a model’s training. In Each fold, data is split into train and test. The model is trained on train data and tested on test data.
CNN is a feedforward neural network [] with a deep structure and convolutional computation. One of the most well-known deep learning algorithms is this one because it performs translation invariant classification, also known as SIANN. As seen in Figure 1, feature extraction is performed by combining the input with convolutional kernels. The pooling layer reduces the network’s computational volume without affecting the feature map’s resolution. The size of the pooling layers in CNNs usually decreases as the number of layers increases. Max pooling and average pooling are two of the most popular types of pooling layers. Convolutional layers extract image features. Max-pooling layers are commonly applied in CNN designs to minimize the number of features. FC is the last 2D-CNN component, defined in the previous layer by the activation function. The SoftMax feature is usually employed. Sigmond’s activation function works effectively in binary classification tasks in this deep-learning architecture. Combine 2D-CNN Sigmoid for better results with a support vector maker (SVM). Our work used the max pooling layer for downsampling and the ReLU activation function. We perform experiments with different filter sizes and hidden layer neurons. We used Adam Optimizer to optimize the weights of the network. Finally, the output is produced using the SoftMax activation function.
We used CNN+SVM, CNN+RF, and CNN+XGBOOST to classify pneumonia and COVID-19. The section below discusses the details of SVM, RF, and XGBOOST. We used the hybrid approach to identify how accurately traditional machine learning models work with the deep learning extracted features. The features are extracted using the CNN model and passed to traditional machine learning models such as RF, SVM, and xgboost.
The SVM approach [], based on statistical learning theory (SLT), has made considerable advancements in machine learning in recent years. The kernel function is the most important element of SVM. By setting a linear boundary in a high-dimensional space and transforming the optimization challenge into a core issue, the kernel function can solve the nonlinear problem in the number of input feature vectors. RF [] is a tree-based ensemble model used for classification and regression problems. It constructs multiple trees of decision trees and outputs the class that is the mode of classes. Xtreme Gradient Boosting [] is an ensemble learning technique that combines weak learners and improves prediction accuracy. It can be used for both classification and regression problems. In our work, we trained the XGBOOST model for COVID-19, pneumonia, and detection on the output first dense layer of CNN.
This study used the accuracy, precision, and recall metrics to validate a machine learning model. The accuracy indicates how accurately our model is predicting the classes. The precision metrics refer to how acutely our model predicts the positive courses. While the recall metrics refer to how accurately the model predicts the positive classes. Utilizing these metrics helps the stakeholders accurately validate a learning model’s performance.
Pneumonia and COVID-19 are severe diseases that affect the lungs of the human body. In our work, we performed experiments on the Pneumonia and COVID-19 datasets to identify pneumonia and COVID-19 patients. We used CNN, the Hybrid model (CNN+SVM, CNN+RF, CNN+xgboost), and different pretrained models. We also compared optimizers such as RMS Prop, stochastic gradient descent, and Adam optimizer. Experimental findings conclude that Adam performs well compared to RMS Prop, with stochastic gradient descent not stuck in local optima. However, the Adam optimizer learns slowly as compared to another optimizer due to high computation in updating weights.
Table 1 shows CNN’s results in pneumonia classification. CNN’s recall is 0.9947, and its accuracy is 0.9756 on the validation set. Recall shows that the false negative rate is low, and this method detects pneumonia with a high detection rate.
Figure 3 shows the pneumonia and normal X-ray pictures with their predicted and actual class label. It demonstrates that this model predicts results are close to the actual label.
Table 2 shows the results of a hybrid approach. CNN+RF achieved the highest validation performance in pneumonia detection as compared to other hybrid methods.
Table 3 shows the results of CNN and SVM with PCA for COVID-19, pneumonia, and regular patients. CNN has the highest performance accuracy compared to SVM with PCA. CNN’s performance is better because feature learning is performed using a convolutional layer, and it detects more visual features than PCA. However, some critical information may be lost during image dimensionality reduction.
This study utilized CNN and a hybrid (CNN traditional machine learning model) to predict pneumonia and COVID-19. The experimental findings show that the stand-alone CNN performs better than the hybrid models. The intuition behind the better results of CNN is that it performs end-to-end learning. On the other hand, the hybrid model can cause overfitting due to additional parameters and complexity.
COVID-19 and Pneumonia are dangerous diseases. Detection and diagnosis of this disease are necessary before they affect the lungs of the human body. Machine Learning and deep learning methods have wide applications in medical images. Our work used CNN and hybrid methods to detect Pneumonia and COVID-19. We performed experiments on two Kaggle datasets of Pneumonia and COVID-19. The experimental result shows that CNN detected pneumonia with a 99.47% recall, which is the highest rate of recall compared to other methods applied in the pneumonia dataset. CNN also achieved the highest results as compared to SVM+PCA on COVID-19 and pneumonia’s combined dataset. We also apply some pretrained models on both datasets. The Extensive experiment in the training of CNN concludes that Adam optimizer performs well compared to RMS Prop, Stochastic gradient descent. In the future, we will increase the performance of these datasets by training CNN with different parameters and using a weighted voting ensemble classifier as a hybrid method.
This study utilized publicly available data. https://www.kaggle.com/datasets/prashant268/chest-xray-COVID-19-pneumonia.
Aristanti S. The analysis of directive speech acts on World Health Organization's speech entitled "WHO Director General's opening remarks at the media briefing on COVID-19-11 May 2020". [dissertation]. UIN SMH Banten; 2021. Available from: https://repository.uinbanten.ac.id/6726/
Birman D. Investigation of the effects of COVID-19 on different organs of the body. Eurasian J Chem Med Pet Res. 2023;2(1):24-36. Available from: https://www.ejcmpr.com/article_160994.html
Tuncer T, Ozyurt F, Dogan S, Subasi A. A novel Covid-19 and pneumonia classification method based on F-transform. Chemometr Intell Lab Syst. 2021;210:104256. Available from: https://pubmed.ncbi.nlm.nih.gov/33531722/
Field EL, Rodriguez AJ, Rajan M, Smith K. Efficacy of artificial intelligence in the categorisation of pediatric pneumonia on chest radiographs: a systematic review. Children (Basel). 2023;10(3):576. Available from: https://pubmed.ncbi.nlm.nih.gov/36980134/
Irmici G, Cè M, Caloro E, Khenkina N, Della Pepa G, Ascenti V, et al. Chest X-ray in Emergency Radiology: What Artificial Intelligence Applications Are Available? Diagnostics (Basel). 2023;13(2):216. Available from: https://pubmed.ncbi.nlm.nih.gov/36673027/
Ukwuoma CC, Qin Z, Belal Bin Heyat M, Akhtar F, Bamisile O, Muaad AY, et al. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest X-ray images. J Adv Res. 2023;48:191-211. Available from: https://pubmed.ncbi.nlm.nih.gov/36084812/
Ren H, Fengshi Jing, Zhurong Chen, Shan He, Jiandong Zhou, Le Liu, et al. CheXMed: a multimodal learning algorithm for pneumonia detection in the elderly. Inf Sci (Ny). 2024;654:119854. Available from: https://www.sciencedirect.com/science/article/abs/pii/S0020025523014391
Malik H, Anees T, Din M, Naeem A. CDC_Net: multi-classification convolutional neural network model for detection of COVID-19, pneumothorax, pneumonia, lung Cancer, and tuberculosis using chest X-rays. Multimed Tools Appl. 2023;82(9):13855-13880. Available from: https://pubmed.ncbi.nlm.nih.gov/36157356/
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233473/
Zhang J, Xie Y, Pang G, Liao Z, Verjans J, Li W, Sun Z, He J, Li Y, Shen C, Xia Y. Viral Pneumonia Screening on Chest X-Rays Using Confidence-Aware Anomaly Detection. IEEE Trans Med Imaging. 2021;40(3):879-890. Available from: https://pubmed.ncbi.nlm.nih.gov/33245693/
Ullah Z, Muhammad Usman, Siddique Latif, Jeonghwan Gwak. Densely attention mechanism based network for COVID-19 detection in chest X-rays. Sci Rep. 2023;13(1):261. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9816547/
Hemdan EE, Shouman MA, Karar ME. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv Preprint. 2020. Available from: https://arxiv.org/abs/2003.11055
Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24:1207-1220. Available from: https://pubmed.ncbi.nlm.nih.gov/33994847/
Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425-434. Available from: https://www.thelancet.com/article/S1473-3099(20)30086-4/fulltext
Toğaçar M, Ergen B, Cömert Z, Özyurt F. A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM. 2020;41(4):212-222. Available from: https://www.sciencedirect.com/science/article/abs/pii/S1959031819301174
Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed. 2020;187:104964. Available from: https://pubmed.ncbi.nlm.nih.gov/31262537/
Jaiswal AK, Tiwari P, Kumar S, Gupta D, Khanna A, Rodrigues JJ. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement. 2019;145:511-518. Available from: https://uobrep.openrepository.com/handle/10547/623797
Bandyopadhyay SK, Dutta S. Machine learning approach for confirmation of COVID-19 cases: positive, negative, death and release (preprint). 2020;172-177.
Wang L, Lin ZQ, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):19549. Available from: https://pubmed.ncbi.nlm.nih.gov/33177550/
Sethy PK, Behera SK. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020; 2020030300. Available from: https://www.preprints.org/manuscript/202003.0300/v1
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. Available from: https://pubmed.ncbi.nlm.nih.gov/32568675/
Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635-640. Available from: https://pubmed.ncbi.nlm.nih.gov/32524445/
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE ACM Trans Comput Biol Bioinform. 2021;18(6):2775-2780. Available from: https://pubmed.ncbi.nlm.nih.gov/33705321/
Bougourzi F, Fadi Dornaika, Cosimo Distante, Abdelmalik Taleb-Ahmed. Emb-trattunet: a novel edge loss function and transformer-CNN architecture for multi-classes pneumonia infection segmentation in low annotation regimes. Artif Intell Rev. 2024;57(4):90. Available from: https://hal.science/hal-04520691v1
Guddanti SS, Apurva Padhye, Anil Prabhakar, Sridhar Tayur. Pneumonia detection by binary classification: classical, quantum, and hybrid approaches for support vector machine (SVM). Front Comput Sci. 2024;5:1286657. Available from: https://www.frontiersin.org/journals/computer-science/articles/10.3389/fcomp.2023.1286657/full
Wang Y, Liu ZL, Yang H, Li R, Liao SJ, Huang Y, et al. Prediction of viral pneumonia based on machine learning models analyzing pulmonary inflammation index scores. Comput Biol Med. 2024;169:107905. Available from: https://pubmed.ncbi.nlm.nih.gov/38159398/
Li X, Xiong X, Liang Z, Tang Y. A machine learning diagnostic model for Pneumocystis jirovecii pneumonia in patients with severe pneumonia. Intern Emerg Med. 2023;18(6):1741-1749. Available from: https://pubmed.ncbi.nlm.nih.gov/37530943/
Khan QA. A Machine Learning-based Method for COVID-19 and Pneumonia Detection. IgMin Res. Jul 05, 2024; 2(7): 518-523. IgMin ID: igmin211; DOI:10.61927/igmin211; Available at: igmin.link/p211
次のリンクを共有した人は、このコンテンツを読むことができます:
Address Correspondence:
Qazi Waqas Khan, Department of Computer Engineering, Jeju National University, Jejusi 63243, Jeju Special Self-Governing Province, Republic of Korea, Email: [email protected]
How to cite this article:
Khan QA. A Machine Learning-based Method for COVID-19 and Pneumonia Detection. IgMin Res. Jul 05, 2024; 2(7): 518-523. IgMin ID: igmin211; DOI:10.61927/igmin211; Available at: igmin.link/p211
Copyright: © 2024 Khan QW This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
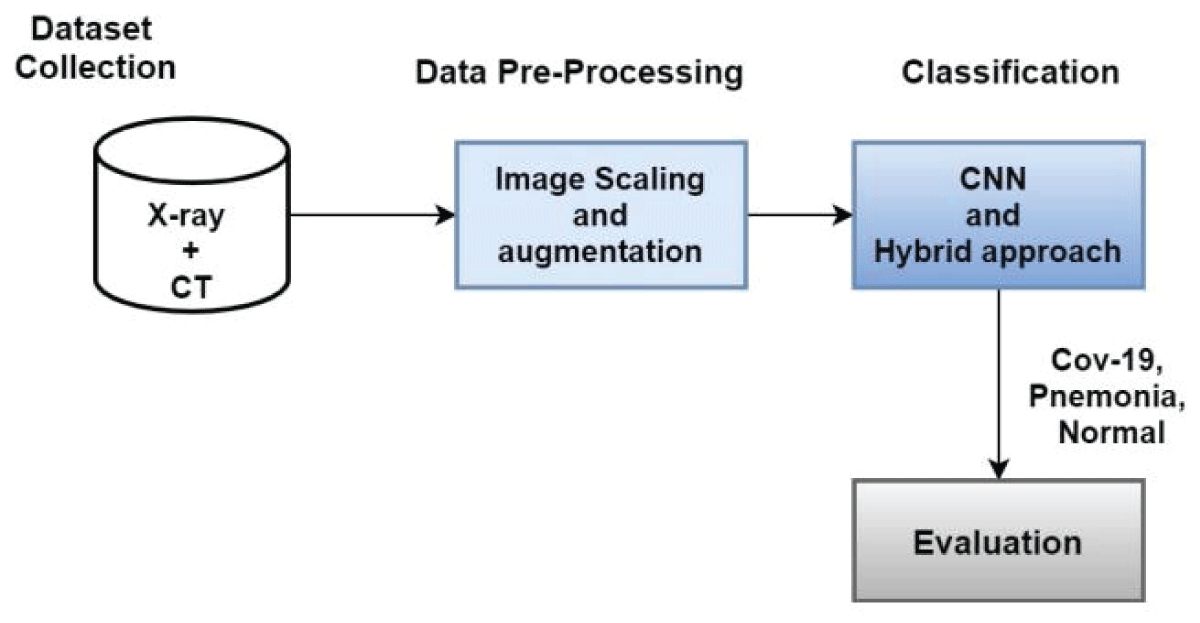 Figure 1: Proposed Methodology Diagram....
Figure 1: Proposed Methodology Diagram....
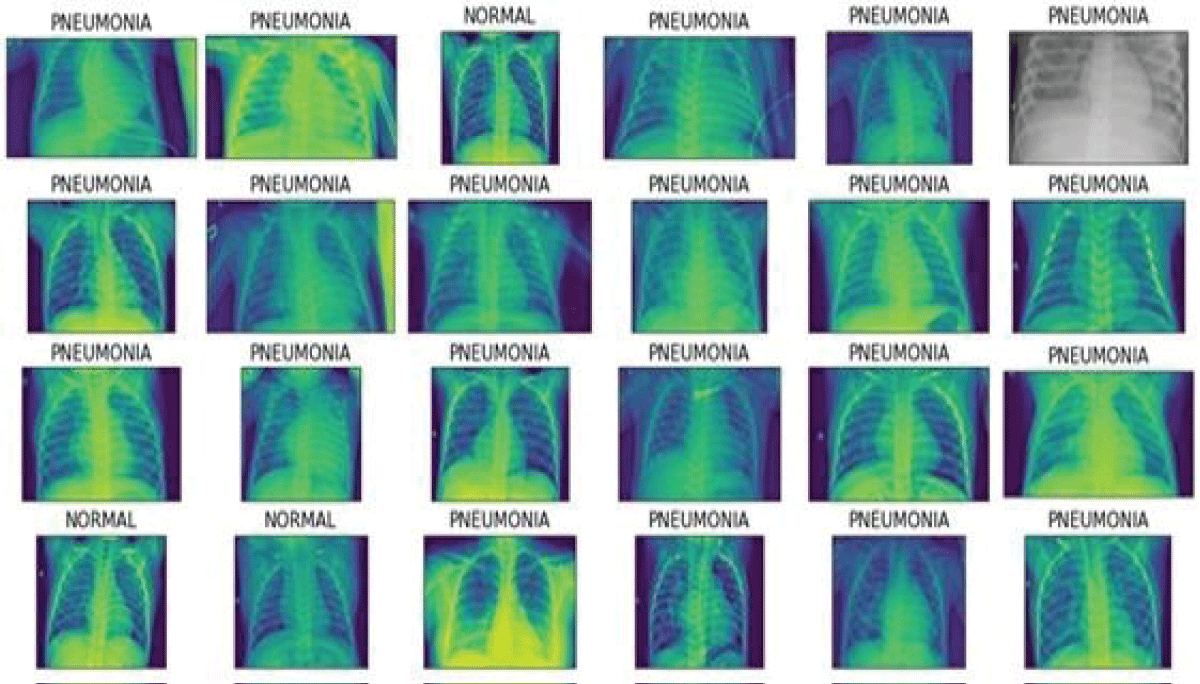 Figure 2: Pneumonia images....
Figure 2: Pneumonia images....
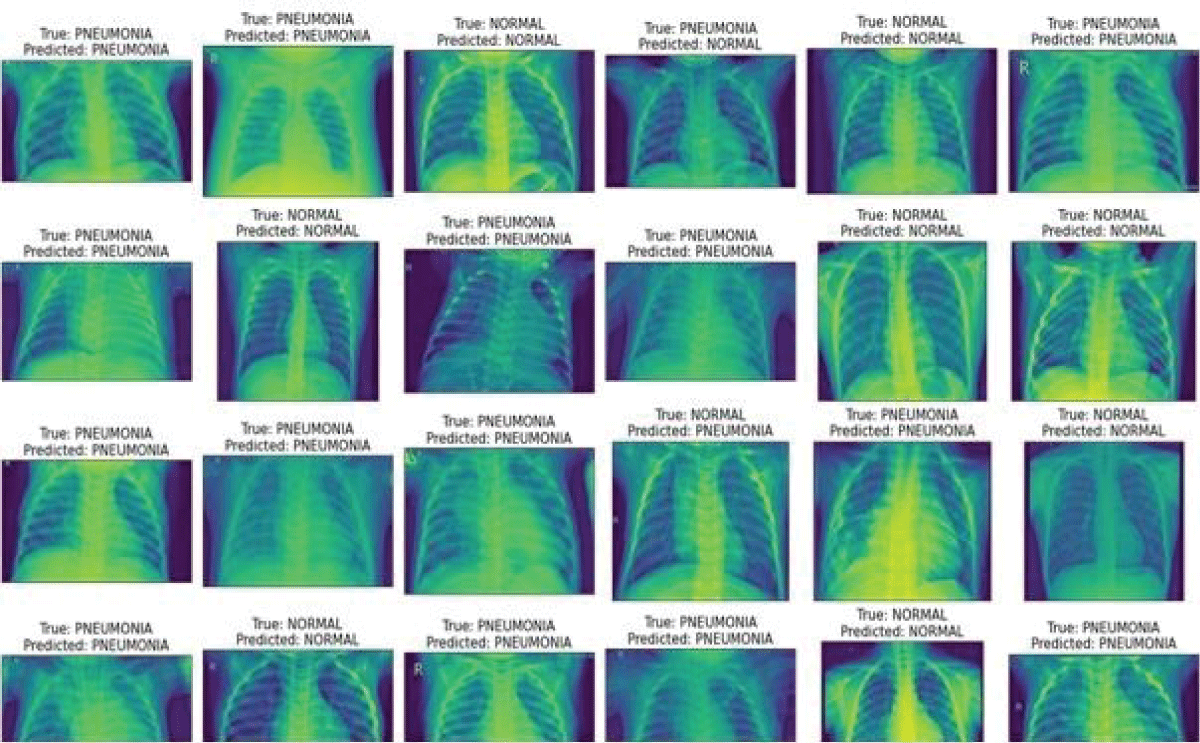 Figure 3: Actual and predicted label images....
Figure 3: Actual and predicted label images....
 Table 1: Results Table of Convolutional Neural Network....
Table 1: Results Table of Convolutional Neural Network....
 Table 2: Results Table of Hybrid model for Pneumonia classi...
Table 2: Results Table of Hybrid model for Pneumonia classi...
 Table 3: Results Comparison on COVID-19 dataset....
Table 3: Results Comparison on COVID-19 dataset....
Aristanti S. The analysis of directive speech acts on World Health Organization's speech entitled "WHO Director General's opening remarks at the media briefing on COVID-19-11 May 2020". [dissertation]. UIN SMH Banten; 2021. Available from: https://repository.uinbanten.ac.id/6726/
Birman D. Investigation of the effects of COVID-19 on different organs of the body. Eurasian J Chem Med Pet Res. 2023;2(1):24-36. Available from: https://www.ejcmpr.com/article_160994.html
Tuncer T, Ozyurt F, Dogan S, Subasi A. A novel Covid-19 and pneumonia classification method based on F-transform. Chemometr Intell Lab Syst. 2021;210:104256. Available from: https://pubmed.ncbi.nlm.nih.gov/33531722/
Field EL, Rodriguez AJ, Rajan M, Smith K. Efficacy of artificial intelligence in the categorisation of pediatric pneumonia on chest radiographs: a systematic review. Children (Basel). 2023;10(3):576. Available from: https://pubmed.ncbi.nlm.nih.gov/36980134/
Irmici G, Cè M, Caloro E, Khenkina N, Della Pepa G, Ascenti V, et al. Chest X-ray in Emergency Radiology: What Artificial Intelligence Applications Are Available? Diagnostics (Basel). 2023;13(2):216. Available from: https://pubmed.ncbi.nlm.nih.gov/36673027/
Ukwuoma CC, Qin Z, Belal Bin Heyat M, Akhtar F, Bamisile O, Muaad AY, et al. A hybrid explainable ensemble transformer encoder for pneumonia identification from chest X-ray images. J Adv Res. 2023;48:191-211. Available from: https://pubmed.ncbi.nlm.nih.gov/36084812/
Ren H, Fengshi Jing, Zhurong Chen, Shan He, Jiandong Zhou, Le Liu, et al. CheXMed: a multimodal learning algorithm for pneumonia detection in the elderly. Inf Sci (Ny). 2024;654:119854. Available from: https://www.sciencedirect.com/science/article/abs/pii/S0020025523014391
Malik H, Anees T, Din M, Naeem A. CDC_Net: multi-classification convolutional neural network model for detection of COVID-19, pneumothorax, pneumonia, lung Cancer, and tuberculosis using chest X-rays. Multimed Tools Appl. 2023;82(9):13855-13880. Available from: https://pubmed.ncbi.nlm.nih.gov/36157356/
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233473/
Zhang J, Xie Y, Pang G, Liao Z, Verjans J, Li W, Sun Z, He J, Li Y, Shen C, Xia Y. Viral Pneumonia Screening on Chest X-Rays Using Confidence-Aware Anomaly Detection. IEEE Trans Med Imaging. 2021;40(3):879-890. Available from: https://pubmed.ncbi.nlm.nih.gov/33245693/
Ullah Z, Muhammad Usman, Siddique Latif, Jeonghwan Gwak. Densely attention mechanism based network for COVID-19 detection in chest X-rays. Sci Rep. 2023;13(1):261. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9816547/
Hemdan EE, Shouman MA, Karar ME. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv Preprint. 2020. Available from: https://arxiv.org/abs/2003.11055
Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021;24:1207-1220. Available from: https://pubmed.ncbi.nlm.nih.gov/33994847/
Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425-434. Available from: https://www.thelancet.com/article/S1473-3099(20)30086-4/fulltext
Toğaçar M, Ergen B, Cömert Z, Özyurt F. A deep feature learning model for pneumonia detection applying a combination of mRMR feature selection and machine learning models. IRBM. 2020;41(4):212-222. Available from: https://www.sciencedirect.com/science/article/abs/pii/S1959031819301174
Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed. 2020;187:104964. Available from: https://pubmed.ncbi.nlm.nih.gov/31262537/
Jaiswal AK, Tiwari P, Kumar S, Gupta D, Khanna A, Rodrigues JJ. Identifying pneumonia in chest X-rays: a deep learning approach. Measurement. 2019;145:511-518. Available from: https://uobrep.openrepository.com/handle/10547/623797
Bandyopadhyay SK, Dutta S. Machine learning approach for confirmation of COVID-19 cases: positive, negative, death and release (preprint). 2020;172-177.
Wang L, Lin ZQ, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):19549. Available from: https://pubmed.ncbi.nlm.nih.gov/33177550/
Sethy PK, Behera SK. Detection of coronavirus disease (COVID-19) based on deep features. Preprints. 2020; 2020030300. Available from: https://www.preprints.org/manuscript/202003.0300/v1
Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. Available from: https://pubmed.ncbi.nlm.nih.gov/32568675/
Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635-640. Available from: https://pubmed.ncbi.nlm.nih.gov/32524445/
Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE ACM Trans Comput Biol Bioinform. 2021;18(6):2775-2780. Available from: https://pubmed.ncbi.nlm.nih.gov/33705321/
Bougourzi F, Fadi Dornaika, Cosimo Distante, Abdelmalik Taleb-Ahmed. Emb-trattunet: a novel edge loss function and transformer-CNN architecture for multi-classes pneumonia infection segmentation in low annotation regimes. Artif Intell Rev. 2024;57(4):90. Available from: https://hal.science/hal-04520691v1
Guddanti SS, Apurva Padhye, Anil Prabhakar, Sridhar Tayur. Pneumonia detection by binary classification: classical, quantum, and hybrid approaches for support vector machine (SVM). Front Comput Sci. 2024;5:1286657. Available from: https://www.frontiersin.org/journals/computer-science/articles/10.3389/fcomp.2023.1286657/full
Wang Y, Liu ZL, Yang H, Li R, Liao SJ, Huang Y, et al. Prediction of viral pneumonia based on machine learning models analyzing pulmonary inflammation index scores. Comput Biol Med. 2024;169:107905. Available from: https://pubmed.ncbi.nlm.nih.gov/38159398/
Li X, Xiong X, Liang Z, Tang Y. A machine learning diagnostic model for Pneumocystis jirovecii pneumonia in patients with severe pneumonia. Intern Emerg Med. 2023;18(6):1741-1749. Available from: https://pubmed.ncbi.nlm.nih.gov/37530943/