
Enforcement and Enlargement of the Saccharomyces cerevisiae Endoplasmic Reticulum through Artificial Evocation of the Unfolded Protein Response
Molecular Biology BiotechnologyMicrobiology受け取った 11 Jan 2024 受け入れられた 29 Jan 2024 オンラインで公開された 30 Jan 2024
Focusing on Biology, Medicine and Engineering ISSN: 2995-8067 | Quick Google Scholar


受け取った 11 Jan 2024 受け入れられた 29 Jan 2024 オンラインで公開された 30 Jan 2024
Upon dysfunction of the Endoplasmic Reticulum (ER), eukaryotic cells provoke a gene expression program, namely, the Unfolded Protein Response (UPR), leading to an increase in the size and function of the ER. In the yeast Saccharomyces cerevisiae, the UPR is modulated by the Hac1i protein, which is a transcription factor produced by ER stress. When the UPR is artificially triggered under non-stress conditions by artificial expression of the Hac1i protein, S. cerevisiae cells carry an enforced and enlarged ER, which allows us to obtain commercially valuable materials such as secretory proteins and functional lipids abundantly.
Owing to their rapid and cost-effective growth characteristics, various species of yeast are frequently employed as platforms for the production of commercially valuable substances. Saccharomyces cerevisiae, also known as baker’s yeast, is used not only for ethanol fermentation but also for basic and applied molecular biology, partly because a large number of genetic engineering techniques have been developed for this organism. A notable example of yeast application lies in the realm of production of human secretory proteins such as antibodies and interferons, which are widely used as biopharmaceutical agents []. Moreover, yeasts can potentially serve as catalysts for the production of lipidic compounds. Through the utilization of genetically modified yeasts, it is now possible to synthesize plant-derived functional lipids [].
The cellular compartment known as the Endoplasmic Reticulum (ER) is the site at which these molecules are synthesized through various biological reactions. In the ER, secretory proteins undergo complicated processes of folding and modification processes, involving the acquisition of cysteine disulfide bonds and N-linked glycosylation chains, which are essential for their subsequent transport to the cell surface. Additionally, a number of enzymes that facilitate lipid biosynthesis are located in the ER membrane. In the ER, lipid droplets, which act as storage sites for neutral lipids, such as triglycerides, emerge as budding structures [,]. The ER in higher eukaryotes also serves as a site in which calcium ions are stored. On the other hand, the calcium ion concentration in the yeast ER is reportedly fairly low [].
The ER, a sac surrounded by a single biological membrane, can take either flat or tubular shapes. The tubular ER often undergoes branching and fusion processes, resulting in the formation of a network-like structure. Notably, the ER occupies a significant portion of the cytoplasm in a wide variety of animal and plant cells. However, in yeast cells, the ER seems to be less developed, forming only two distinct regions: the peripheral ER and the inner ER []. The peripheral ER, which can be found adjacent to the plasma membrane, is composed of a complex network of interconnected tubules within yeast cells []. On the other hand, the inner ER is an alias of the nuclear envelope, a structure that envelops chromosomal DNA, thus forming the nucleus. Therefore, it is reasonable to hypothesize that by expanding this organelle in yeast cells, the capacity of the yeast cells to generate valuable substances can be considerably enhanced.
Applying the Unfolded Protein Response (UPR) is a possible method for enhancing the size and function of the ER. ER stress, which is triggered by dysfunction or deficiency in ER function, is often accompanied by the accumulation of unfolded proteins in the ER. Eukaryotic cells commonly have the ability to provoke a protective response known as the Unfolded Protein Response (UPR) to counteract ER stress and ensure the robustness of the ER [].
The molecular mechanism regulating the UPR was initially elucidated through pioneering investigations using S. cerevisiae as an exemplary model organism []. Ire1, a transmembrane ribonuclease located in the ER, is evolutionarily conserved across various eukaryotic species and serves as a sensor of ER stress. In fungal species belonging to the phylum Ascomycetes, which includes S. cerevisiae, Ire1 facilitates the splicing of the HAC1 gene transcript in response to ER stress (Figure 1). In S. cerevisiae cells, the unspliced variant of HAC1 mRNA (referred to as HAC1u, with the letter u representing “uninduced”) exhibits poor translational efficiency. Moreover, the Hac1u protein is extremely unstable. Conversely, the spliced form of HAC1 mRNA (designated HAC1i, with the letter i representing “induced”) undergoes translation to yield a transcription factor known as the Hac1i protein, which plays the main role in the transcriptional induction on the UPR (Figure 1).
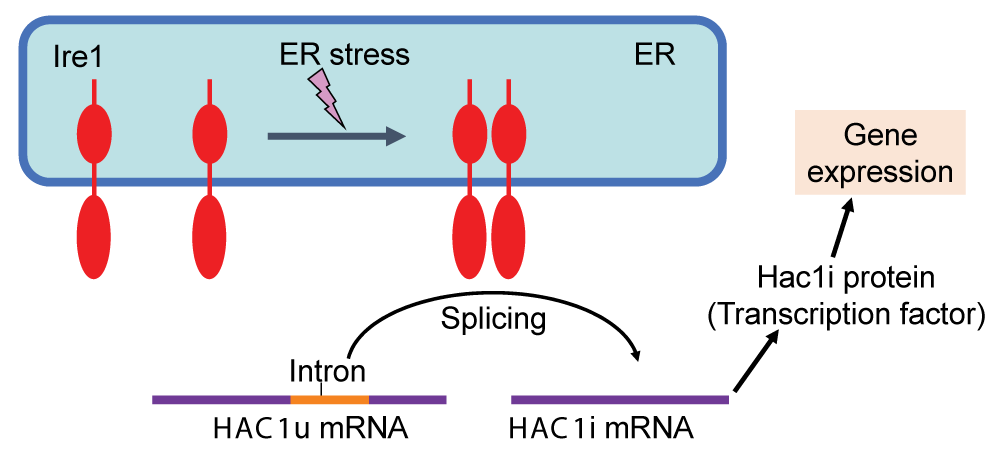 Figure 1: The UPR in S. cerevisiae cells Under non-stress conditions, the HAC1 mRNA (HAC1u) contains the intron sequence and is functionless. Upon ER stress, Ire1 is homo-associated to splice the HAC1u mRNA, yielding the HAC1i mRNA, which is translated to the Hac1i protein to induce gene expression for the UPR.
Figure 1: The UPR in S. cerevisiae cells Under non-stress conditions, the HAC1 mRNA (HAC1u) contains the intron sequence and is functionless. Upon ER stress, Ire1 is homo-associated to splice the HAC1u mRNA, yielding the HAC1i mRNA, which is translated to the Hac1i protein to induce gene expression for the UPR.Transcriptome analyses conducted by our research group and other researchers have provided evidence demonstrating that a number of genes are induced by the UPR in a manner that is controlled by Ire1 and HAC1 in S. cerevisiae cells [-]. The Hac1i-target genes encompass a wide range of functional categories, including those involved in ER-located molecular chaperones and enzymes that facilitate the formation of cysteine disulfide bonds, N-linked sugar chains, and lipidic molecules. Consequently, when the Hac1i protein was expressed in an unregulated manner from a constitutive promoter, protein secretion increased, although the magnitude of this elevation was relatively modest, as demonstrated in the pioneering work of Valkonen, et al. []. More recently, Lin, et al. [] reported that heterologous protein secretion from S. cerevisiae cells was elevated by some genetic mutations that led to an increase in cellular Hac1i protein abundance. The Hac1i protein also increased protein secretion from another yeast species Pichia pastoris [,]. Furthermore, some investigations have revealed that S. cerevisiae cells engineered to artificially express the Hac1i protein exhibit upregulation of the production of lipidic molecules (Figure 2A) [,]. It should also be noted that the ER in S. cerevisiae cells artificially expressing the Hac1i protein is highly expanded (Figure 2B) [].
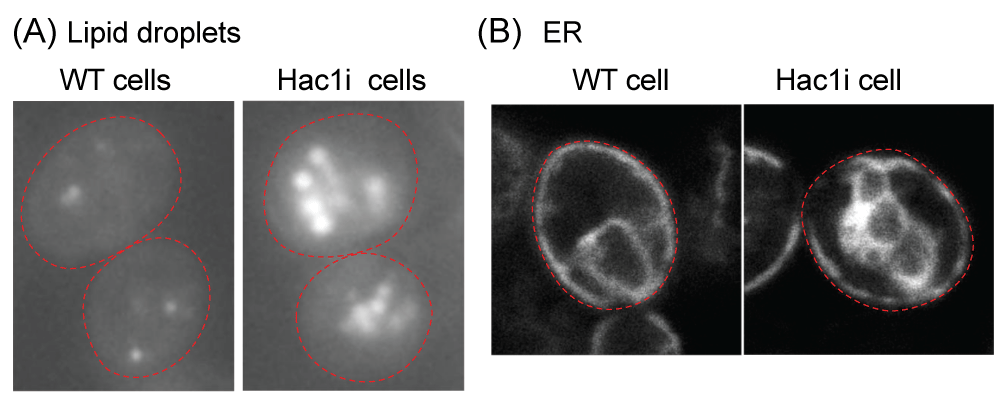 Figure 2: Fluorescence microscopic analysis of S. cerevisiae cells Wild-type S. cerevisiae cells (BY4742 (MATα, ura3, leu2, his3, lys2): WT cell) and their derivative carrying the HAC1i expression plasmid (Hac1i cell) were cultured in yeast standard synthetic complete (SC) medium at 30 °C and fluorescence microscopically observed. To generate the HAC1i expression plasmid, the HAC1i gene was cloned into the Tet-off vector pCM190 [], leading to inducible expression of the Hac1i protein upon culturing cells in the SC medium. (A) Cells were stained with BODIPY 493/503 to visualize neutral lipids. Hac1i cells carried larger and more abundant lipid droplets than WT cells. (B) The fluorescent ER marker protein, Elo2-mCherry, was expressed from a plasmid that had been created by insertion of the ELO2 gene, which encodes an ER membrane protein Elo2, into the mCherry expression plasmid pYT-TDH3p-PMA1-mCherry []. The ER in Hac1i cells was more expanded than in WT cells. Red dashed lines represent the cell outline.
Figure 2: Fluorescence microscopic analysis of S. cerevisiae cells Wild-type S. cerevisiae cells (BY4742 (MATα, ura3, leu2, his3, lys2): WT cell) and their derivative carrying the HAC1i expression plasmid (Hac1i cell) were cultured in yeast standard synthetic complete (SC) medium at 30 °C and fluorescence microscopically observed. To generate the HAC1i expression plasmid, the HAC1i gene was cloned into the Tet-off vector pCM190 [], leading to inducible expression of the Hac1i protein upon culturing cells in the SC medium. (A) Cells were stained with BODIPY 493/503 to visualize neutral lipids. Hac1i cells carried larger and more abundant lipid droplets than WT cells. (B) The fluorescent ER marker protein, Elo2-mCherry, was expressed from a plasmid that had been created by insertion of the ELO2 gene, which encodes an ER membrane protein Elo2, into the mCherry expression plasmid pYT-TDH3p-PMA1-mCherry []. The ER in Hac1i cells was more expanded than in WT cells. Red dashed lines represent the cell outline.However, artificial and constitutive expression of the Hac1i protein has a significant inhibitory effect on the growth of S. cerevisiae cells, as demonstrated by studies conducted by Nguyen, et al. [] and Mori, et al. []. This inhibitory effect leads to various undesired outcomes, such as a reduction and/or delay in the production of biomass, as well as the occurrence of fast-growing mutants with a low UPR. As a result of these issues, achieving stable growth of S. cerevisiae cells constitutively and highly producing the Hac1i protein has become a challenging task.
Nevertheless, in the case of Valkonen, et al. [] and Qu, et al. [], S. cerevisiae cells that continuously produced the Hac1i protein did not exhibit significant retardation of growth. In this particular scenario, it is postulated that the UPR was not potent enough to impede cell growth, presumably due to the low expression level of the Hac1i protein. Indeed, the level of expression of the prominent UPR target gene KAR2, as reported by Valkonen, et al. [], was not as high as that documented by Nguyen, et al. [], where the Hac1i protein was synthesized from the endogenous HAC1 promoter. It is plausible that the growth retardation and the ER productivity resulting from the artificial expression of HAC1i are contradictory issues. In Valkonen, et al. [], when expressed in S. cerevisiae cells, the Hac1i protein derived from Trichoderma reesei led to more severe growth retardation, greater UPR activation, and more secreted protein production than did the authentic S. cerevisiae Hac1i protein.
As a potential solution to this antinomic predicament, we previously developed a methodology wherein ER stress is induced in S. cerevisiae cells expressing the Hac1i protein []. Intriguingly, the retarded growth of S. cerevisiae cells expressing the Hac1i protein was partially ameliorated, albeit without any reduction in the UPR level, when these cells were grown in the presence of weak ER stress stimuli, such as low concentrations of tunicamycin. Using this new methodology, we successfully demonstrated considerable enhancement in the generation of lipidic molecules from S. cerevisiae cells []. Nonetheless, it should also be noted that this technique has a disadvantageous point as ER stress, even at a mild level, can be toxic. In fact, S. cerevisiae cells subjected to mild ER stress exhibited a significant decrease in viability upon continuous culturing.
The ER is a cellular compartment in which a number of materials, some of which are commercially valuable, are biosynthesized. As a biotechnological technique to increase the productivity of the ER, we and others enforced and enlarged the ER through artificial and constative evocation of the UPR, which, in nature, is a cytoprotective response induced upon ER stress in S. cerevisiae cells. One disadvantage of this methodology is that the unregulated UPR induced by the strong and constitutive Hac1i-protein expression severely retards cellular growth.
One possible solution to this problem is to express the Hac1i protein in a regulated manner using an inducible promoter system. Upon the growth phase to yield a large cell mass, the S. cerevisiae cells proliferate rapidly by repressing the Hac1i protein expression. Subsequently, the expression of the Hac1i protein is induced to increase the capacity of the ER to produce desired materials. Another intriguing approach is to identify genes responsible for growth retardation upon artificial induction of the UPR. We anticipate that we can generate rapid-growing and high-UPR yeast strains through genetic modifications to delete such genes.
Nielsen J. Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered. 2013 Jul-Aug;4(4):207-11. doi: 10.4161/bioe.22856. Epub 2012 Nov 12. PMID: 23147168; PMCID: PMC3728191.
Jiang W, Li C, Li Y, Peng H. Metabolic Engineering Strategies for Improved Lipid Production and Cellular Physiological Responses in Yeast Saccharomyces cerevisiae. J Fungi (Basel). 2022 Apr 21;8(5):427. doi: 10.3390/jof8050427. PMID: 35628683; PMCID: PMC9144191.
Wang CW. Lipid droplet dynamics in budding yeast. Cell Mol Life Sci. 2015 Jul;72(14):2677-95. doi: 10.1007/s00018-015-1903-5. Epub 2015 Apr 18. PMID: 25894691.
Chapman KD, Aziz M, Dyer JM, Mullen RT. Mechanisms of lipid droplet biogenesis. Biochem J. 2019 Jul 9;476(13):1929-1942. doi: 10.1042/BCJ20180021. PMID: 31289128.
Strayle J, Pozzan T, Rudolph HK. Steady-state free Ca(2+) in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 1999 Sep 1;18(17):4733-43. doi: 10.1093/emboj/18.17.4733. PMID: 10469652; PMCID: PMC1171546.
Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991 Dec;7(9):891-911. doi: 10.1002/yea.320070902. PMID: 1803815.
Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000 Aug 7;150(3):461-74. doi: 10.1083/jcb.150.3.461. PMID: 10931860; PMCID: PMC2175198.
Karagöz GE, Acosta-Alvear D, Walter P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a033886. doi: 10.1101/cshperspect.a033886. PMID: 30670466; PMCID: PMC6719602.
Ishiwata-Kimata Y, Kimata Y. Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts. J Fungi (Basel). 2023 Oct 5;9(10):989. doi: 10.3390/jof9100989. PMID: 37888245; PMCID: PMC10608004.
Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000 Apr 28;101(3):249-58. doi: 10.1016/s0092-8674(00)80835-1. PMID: 10847680.
Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006 Jan;11(1):59-69. doi: 10.1111/j.1365-2443.2005.00921.x. PMID: 16371132.
Tran DM, Ishiwata-Kimata Y, Mai TC, Kubo M, Kimata Y. The unfolded protein response alongside the diauxic shift of yeast cells and its involvement in mitochondria enlargement. Sci Rep. 2019 Sep 4;9(1):12780. doi: 10.1038/s41598-019-49146-5. PMID: 31484935; PMCID: PMC6726593.
Valkonen M, Penttilä M, Saloheimo M. Effects of inactivation and constitutive expression of the unfolded- protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2003 Apr;69(4):2065-72. doi: 10.1128/AEM.69.4.2065-2072.2003. PMID: 12676684; PMCID: PMC154816.
Lin Y, Feng Y, Zheng L, Zhao M, Huang M. Improved protein production in yeast using cell engineering with genes related to a key factor in the unfolded protein response. Metab Eng. 2023 May;77:152-161. doi: 10.1016/j.ymben.2023.04.004. Epub 2023 Apr 10. PMID: 37044356.
Raschmanová H, Weninger A, Knejzlík Z, Melzoch K, Kovar K. Engineering of the unfolded protein response pathway in Pichia pastoris: enhancing production of secreted recombinant proteins. Appl Microbiol Biotechnol. 2021 Jun;105(11):4397-4414. doi: 10.1007/s00253-021-11336-5. Epub 2021 May 26. PMID: 34037840; PMCID: PMC8195892.
De Groeve M, Laukens B, Schotte P. Optimizing expression of Nanobody® molecules in Pichia pastoris through co-expression of auxiliary proteins under methanol and methanol-free conditions. Microb Cell Fact. 2023 Jul 22;22(1):135. doi: 10.1186/s12934-023-02132-z. PMID: 37481525; PMCID: PMC10362571.
Qu Z, Zhang L, Zhu S, Yuan W, Hang J, Yin D, Tang X, Zheng J, Wang Z, Sun J. Overexpression of the transcription factor HAC1 improves nerolidol production in engineered yeast. Enzyme Microb Technol. 2020 Mar; 134:109485. doi: 10.1016/j.enzmictec.2019.109485. Epub 2019 Dec 2. PMID: 32044032.
Nguyen PTM, Ishiwata-Kimata Y, Kimata Y. Fast-Growing Saccharomyces cerevisiae Cells with a Constitutive Unfolded Protein Response and Their Potential for Lipidic Molecule Production. Appl Environ Microbiol. 2022 Nov 8;88(21):e0108322. doi: 10.1128/aem.01083-22. Epub 2022 Oct 18. PMID: 36255243; PMCID: PMC9642017.
Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4660-5. doi: 10.1073/pnas.050010197. PMID: 10781071; PMCID: PMC18289.
Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997 Jul;13(9):837-48. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. PMID: 9234672.
Mai TC, Ishiwata-Kimata Y, Le QG, Kido H, Kimata Y. Dispersion of Endoplasmic Reticulum-associated Compartments by 4-phenyl Butyric Acid in Yeast Cells. Cell Struct Funct. 2019 Nov 23;44(2):173-182. doi: 10.1247/csf.19023. Epub 2019 Oct 17. PMID: 31619600.
Monguchi M, Kimata Y. Enforcement and Enlargement of the Saccharomyces cerevisiae Endoplasmic Reticulum through Artificial Evocation of the Unfolded Protein Response. IgMin Res. 30 Jan, 2024; 2(1): 036-038. IgMin ID: igmin142; DOI: 10.61927/igmin142; Available at: www.igminresearch.com/articles/pdf/igmin142.pdf
次のリンクを共有した人は、このコンテンツを読むことができます:
Address Correspondence:
Yukio Kimata, Ph.D. Associate Professor, Division of Biological Science, Department of Science and Technology, Nara Institute of Science and Technology, 8916-5 Takayama, Ikoma, Nara 630-0192, Japan, Email: [email protected]
How to cite this article:
Monguchi M, Kimata Y. Enforcement and Enlargement of the Saccharomyces cerevisiae Endoplasmic Reticulum through Artificial Evocation of the Unfolded Protein Response. IgMin Res. 30 Jan, 2024; 2(1): 036-038. IgMin ID: igmin142; DOI: 10.61927/igmin142; Available at: www.igminresearch.com/articles/pdf/igmin142.pdf
Copyright: © 2024 Monguchi M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
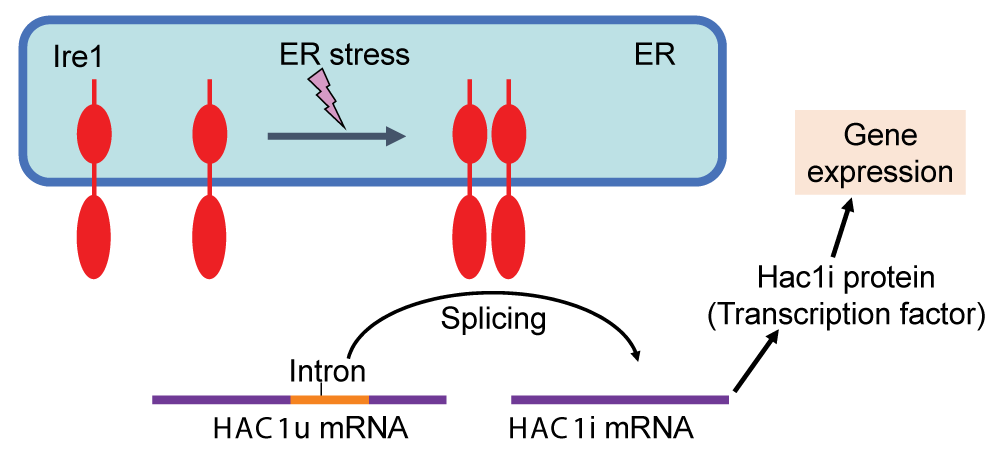 Figure 1: The UPR in S. cerevisiae cells Under non-stress co...
Figure 1: The UPR in S. cerevisiae cells Under non-stress co...
![Fluorescence microscopic analysis of S. cerevisiae cells Wild-type S. cerevisiae cells (BY4742 (MATα, ura3, leu2, his3, lys2): WT cell) and their derivative carrying the HAC1i expression plasmid (Hac1i cell) were cultured in yeast standard synthetic complete (SC) medium at 30 °C and fluorescence microscopically observed. To generate the HAC1i expression plasmid, the HAC1i gene was cloned into the Tet-off vector pCM190 [20], leading to inducible expression of the Hac1i protein upon culturing cells in the SC medium. (A) Cells were stained with BODIPY 493/503 to visualize neutral lipids. Hac1i cells carried larger and more abundant lipid droplets than WT cells. (B) The fluorescent ER marker protein, Elo2-mCherry, was expressed from a plasmid that had been created by insertion of the ELO2 gene, which encodes an ER membrane protein Elo2, into the mCherry expression plasmid pYT-TDH3p-PMA1-mCherry [21]. The ER in Hac1i cells was more expanded than in WT cells. Red dashed lines represent the cell outline.](https://www.igminresearch.jp/articles/figures/igmin142/igmin142.g002.png) Figure 2: Fluorescence microscopic analysis of S. cerevisiae...
Figure 2: Fluorescence microscopic analysis of S. cerevisiae...
Nielsen J. Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered. 2013 Jul-Aug;4(4):207-11. doi: 10.4161/bioe.22856. Epub 2012 Nov 12. PMID: 23147168; PMCID: PMC3728191.
Jiang W, Li C, Li Y, Peng H. Metabolic Engineering Strategies for Improved Lipid Production and Cellular Physiological Responses in Yeast Saccharomyces cerevisiae. J Fungi (Basel). 2022 Apr 21;8(5):427. doi: 10.3390/jof8050427. PMID: 35628683; PMCID: PMC9144191.
Wang CW. Lipid droplet dynamics in budding yeast. Cell Mol Life Sci. 2015 Jul;72(14):2677-95. doi: 10.1007/s00018-015-1903-5. Epub 2015 Apr 18. PMID: 25894691.
Chapman KD, Aziz M, Dyer JM, Mullen RT. Mechanisms of lipid droplet biogenesis. Biochem J. 2019 Jul 9;476(13):1929-1942. doi: 10.1042/BCJ20180021. PMID: 31289128.
Strayle J, Pozzan T, Rudolph HK. Steady-state free Ca(2+) in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 1999 Sep 1;18(17):4733-43. doi: 10.1093/emboj/18.17.4733. PMID: 10469652; PMCID: PMC1171546.
Preuss D, Mulholland J, Kaiser CA, Orlean P, Albright C, Rose MD, Robbins PW, Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991 Dec;7(9):891-911. doi: 10.1002/yea.320070902. PMID: 1803815.
Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000 Aug 7;150(3):461-74. doi: 10.1083/jcb.150.3.461. PMID: 10931860; PMCID: PMC2175198.
Karagöz GE, Acosta-Alvear D, Walter P. The Unfolded Protein Response: Detecting and Responding to Fluctuations in the Protein-Folding Capacity of the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a033886. doi: 10.1101/cshperspect.a033886. PMID: 30670466; PMCID: PMC6719602.
Ishiwata-Kimata Y, Kimata Y. Fundamental and Applicative Aspects of the Unfolded Protein Response in Yeasts. J Fungi (Basel). 2023 Oct 5;9(10):989. doi: 10.3390/jof9100989. PMID: 37888245; PMCID: PMC10608004.
Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000 Apr 28;101(3):249-58. doi: 10.1016/s0092-8674(00)80835-1. PMID: 10847680.
Kimata Y, Ishiwata-Kimata Y, Yamada S, Kohno K. Yeast unfolded protein response pathway regulates expression of genes for anti-oxidative stress and for cell surface proteins. Genes Cells. 2006 Jan;11(1):59-69. doi: 10.1111/j.1365-2443.2005.00921.x. PMID: 16371132.
Tran DM, Ishiwata-Kimata Y, Mai TC, Kubo M, Kimata Y. The unfolded protein response alongside the diauxic shift of yeast cells and its involvement in mitochondria enlargement. Sci Rep. 2019 Sep 4;9(1):12780. doi: 10.1038/s41598-019-49146-5. PMID: 31484935; PMCID: PMC6726593.
Valkonen M, Penttilä M, Saloheimo M. Effects of inactivation and constitutive expression of the unfolded- protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 2003 Apr;69(4):2065-72. doi: 10.1128/AEM.69.4.2065-2072.2003. PMID: 12676684; PMCID: PMC154816.
Lin Y, Feng Y, Zheng L, Zhao M, Huang M. Improved protein production in yeast using cell engineering with genes related to a key factor in the unfolded protein response. Metab Eng. 2023 May;77:152-161. doi: 10.1016/j.ymben.2023.04.004. Epub 2023 Apr 10. PMID: 37044356.
Raschmanová H, Weninger A, Knejzlík Z, Melzoch K, Kovar K. Engineering of the unfolded protein response pathway in Pichia pastoris: enhancing production of secreted recombinant proteins. Appl Microbiol Biotechnol. 2021 Jun;105(11):4397-4414. doi: 10.1007/s00253-021-11336-5. Epub 2021 May 26. PMID: 34037840; PMCID: PMC8195892.
De Groeve M, Laukens B, Schotte P. Optimizing expression of Nanobody® molecules in Pichia pastoris through co-expression of auxiliary proteins under methanol and methanol-free conditions. Microb Cell Fact. 2023 Jul 22;22(1):135. doi: 10.1186/s12934-023-02132-z. PMID: 37481525; PMCID: PMC10362571.
Qu Z, Zhang L, Zhu S, Yuan W, Hang J, Yin D, Tang X, Zheng J, Wang Z, Sun J. Overexpression of the transcription factor HAC1 improves nerolidol production in engineered yeast. Enzyme Microb Technol. 2020 Mar; 134:109485. doi: 10.1016/j.enzmictec.2019.109485. Epub 2019 Dec 2. PMID: 32044032.
Nguyen PTM, Ishiwata-Kimata Y, Kimata Y. Fast-Growing Saccharomyces cerevisiae Cells with a Constitutive Unfolded Protein Response and Their Potential for Lipidic Molecule Production. Appl Environ Microbiol. 2022 Nov 8;88(21):e0108322. doi: 10.1128/aem.01083-22. Epub 2022 Oct 18. PMID: 36255243; PMCID: PMC9642017.
Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4660-5. doi: 10.1073/pnas.050010197. PMID: 10781071; PMCID: PMC18289.
Garí E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997 Jul;13(9):837-48. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. PMID: 9234672.
Mai TC, Ishiwata-Kimata Y, Le QG, Kido H, Kimata Y. Dispersion of Endoplasmic Reticulum-associated Compartments by 4-phenyl Butyric Acid in Yeast Cells. Cell Struct Funct. 2019 Nov 23;44(2):173-182. doi: 10.1247/csf.19023. Epub 2019 Oct 17. PMID: 31619600.