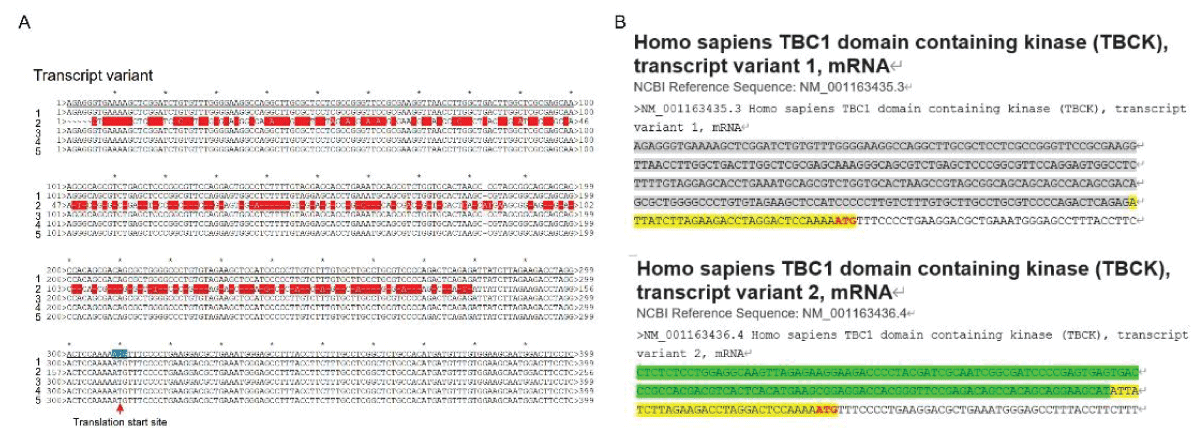
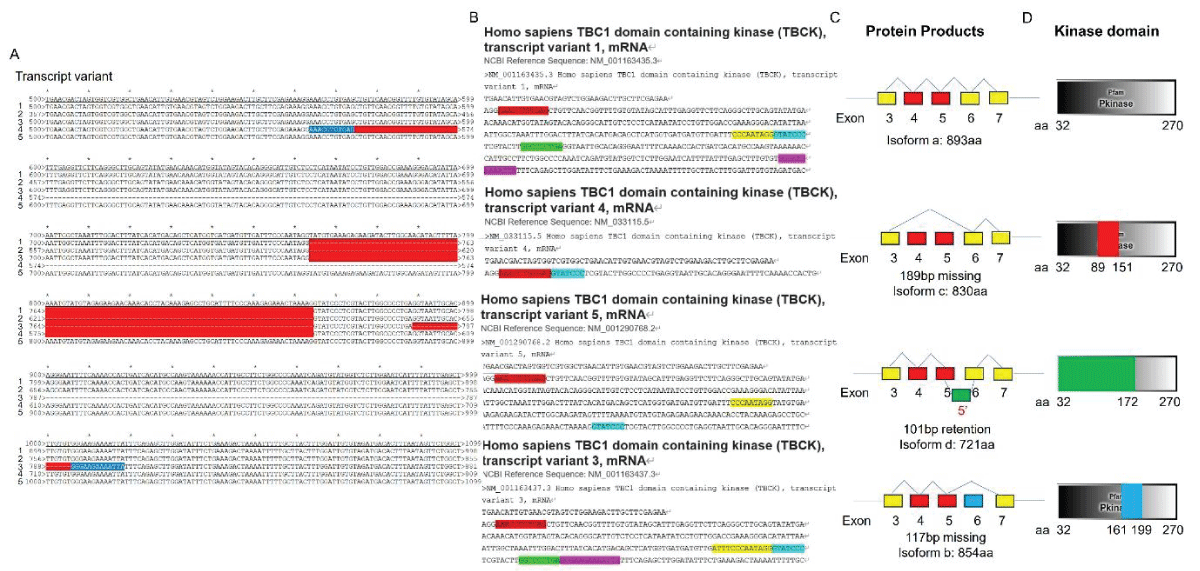
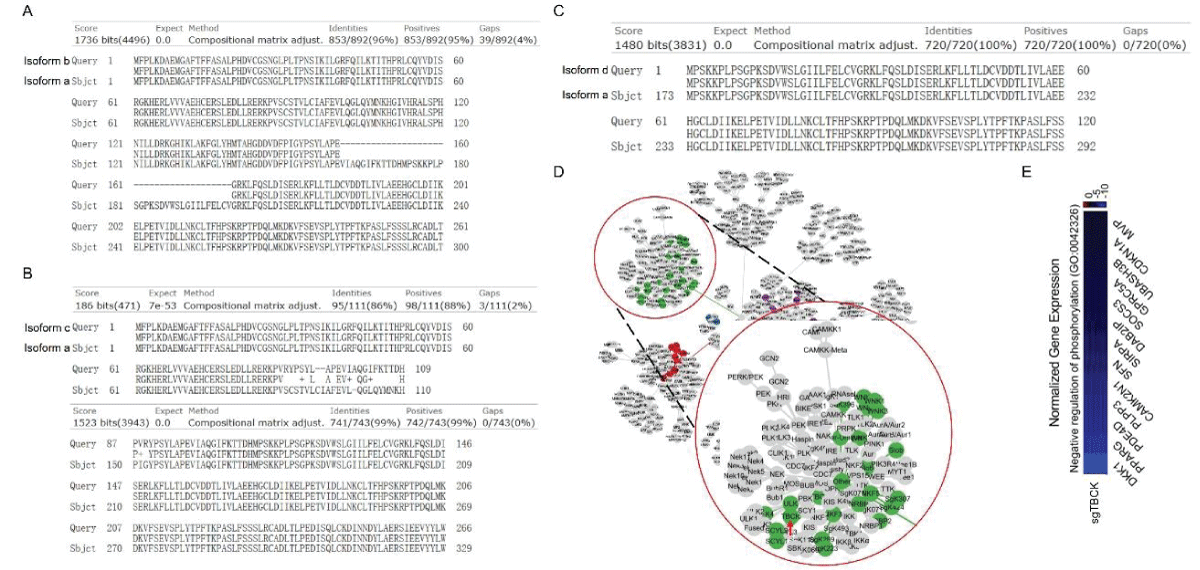
要約
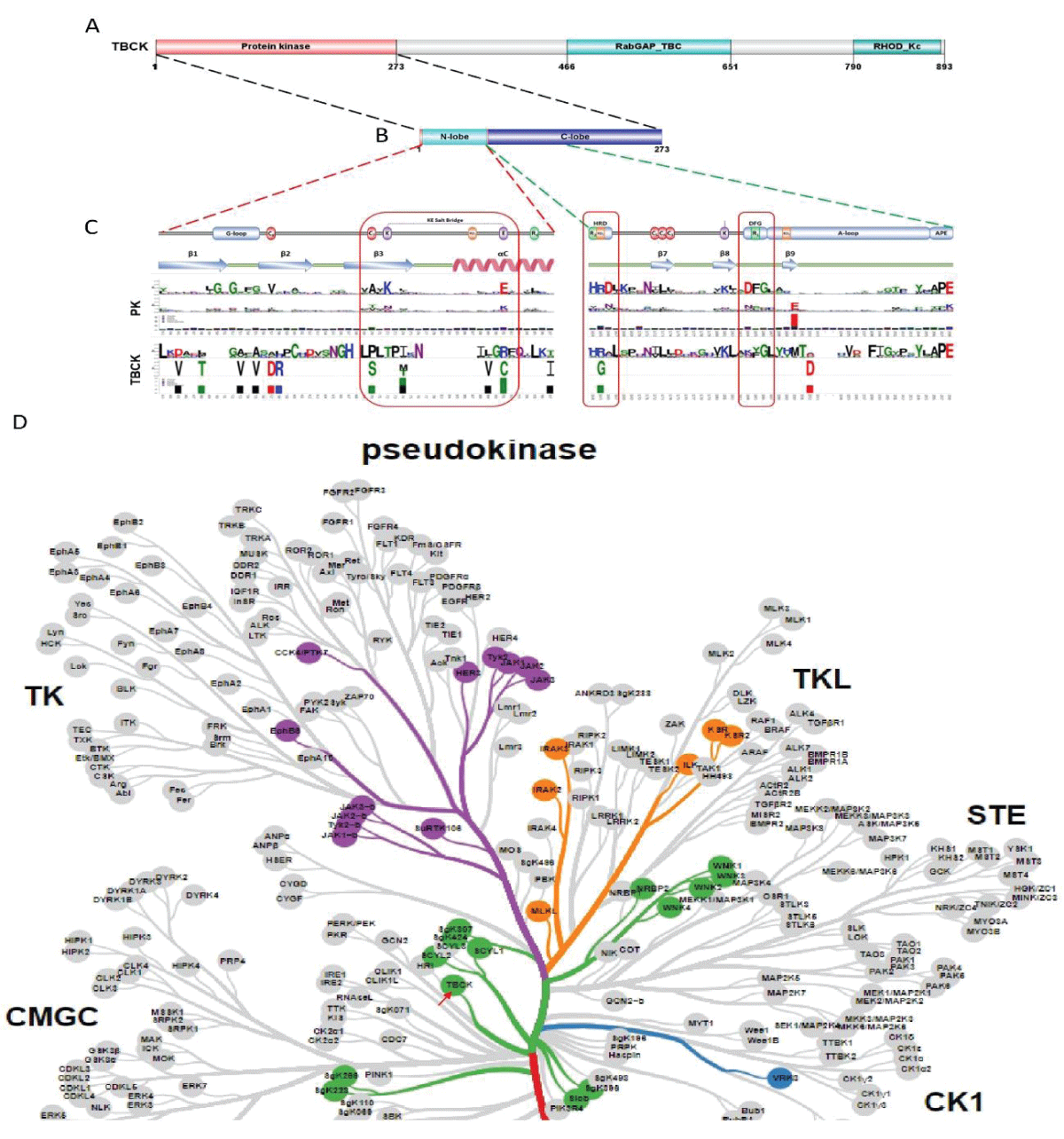
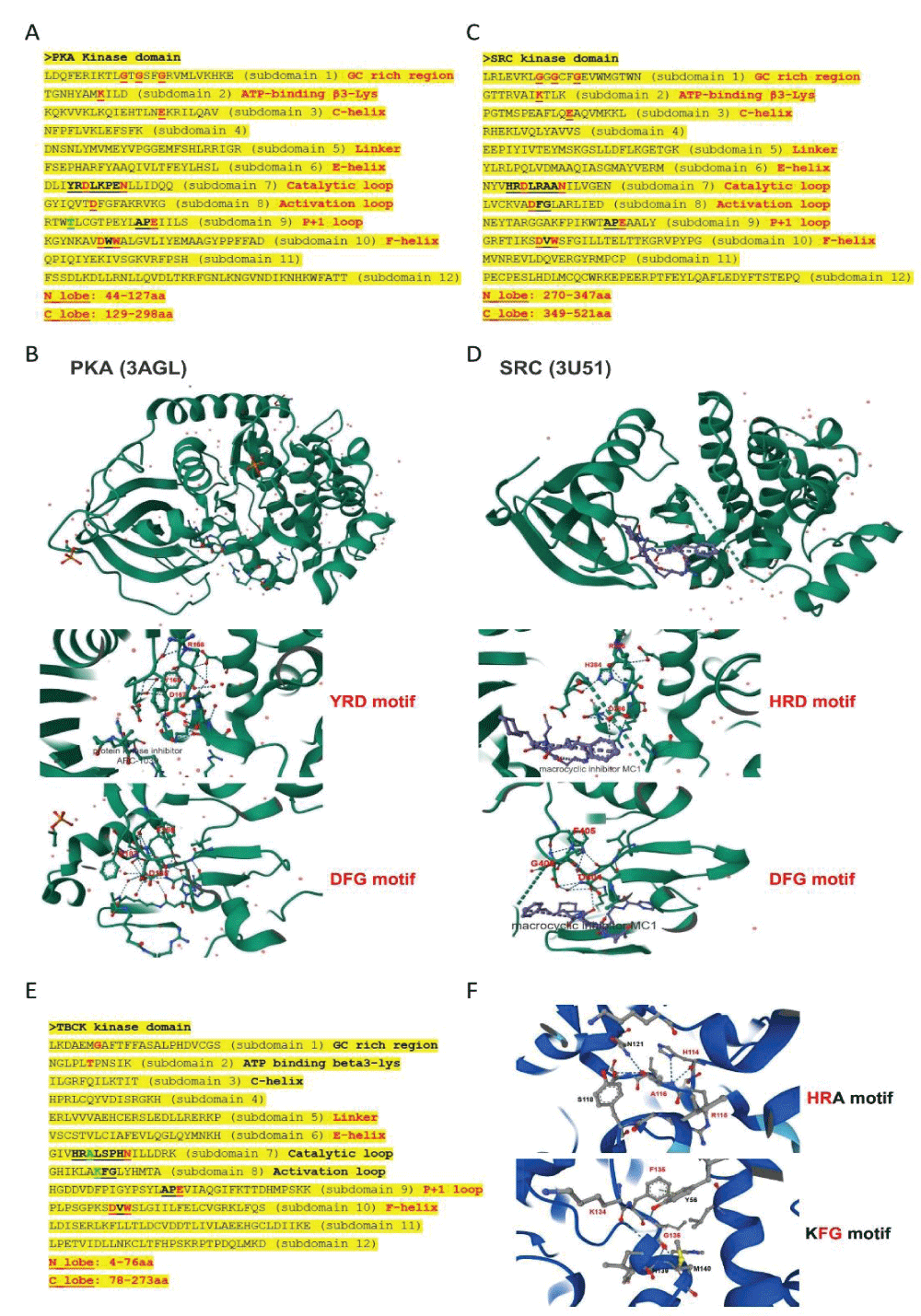
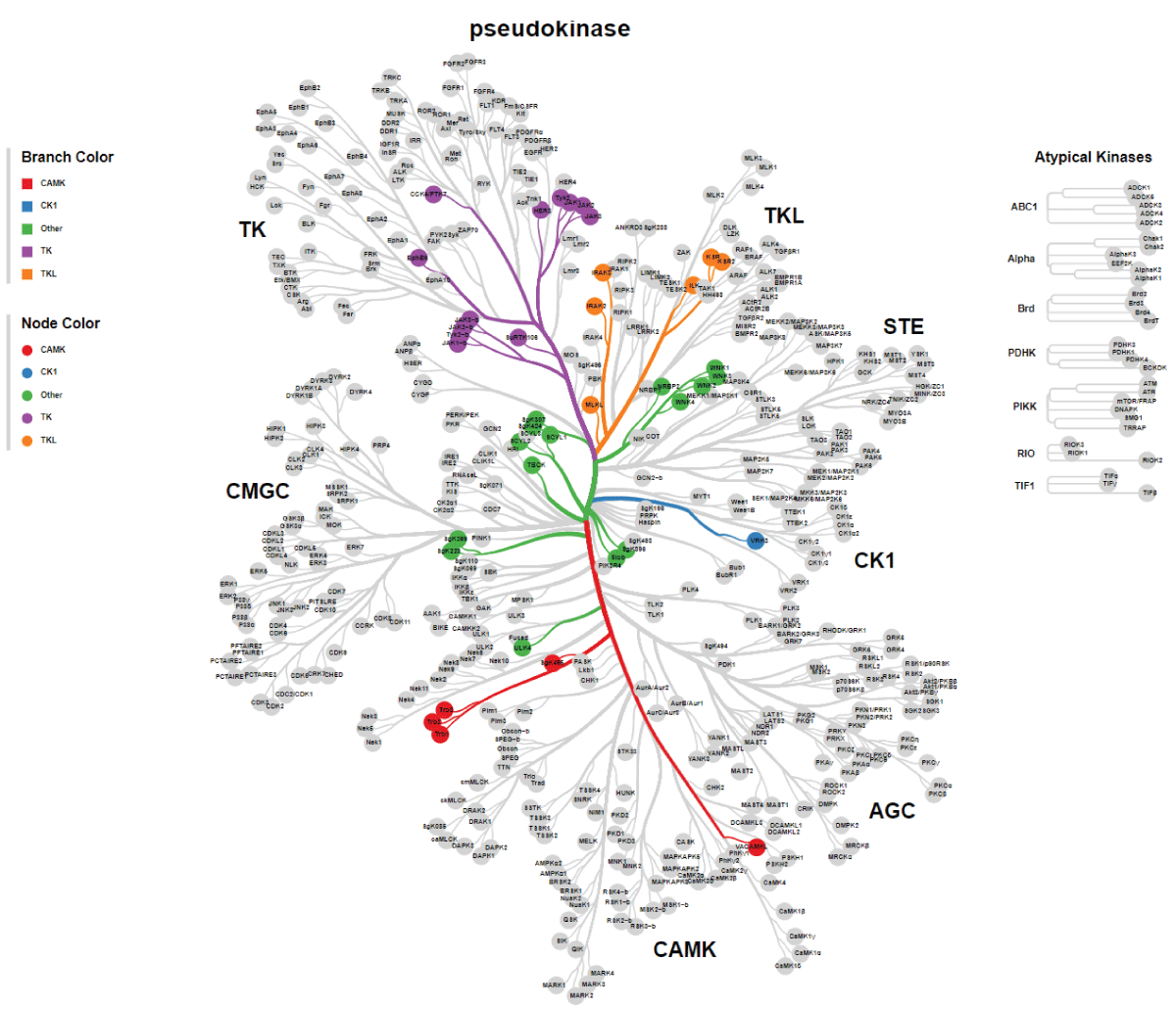
Since the initial identification of TBCK (formerly MGC16169) in 2010, significant advances have been made in understanding the role of TBCK mutations in neurodevelopmental disorders such as TBCK encephalopathy. However, the precise function and detailed mechanisms of TBCK remain largely unexplored. Previous studies, including our own, suggest that aberrant expression or mutations in TBCK can impact cell growth, division, and cytoskeleton assembly, contributing to both cancer and neurogenetic diseases. Despite this, the specific domains within TBCK responsible for these functions are still unclear. Notably, mutations in the TBC domain have been implicated in disrupting mTOR pathways, linking TBCK dysfunction to neurogenetic disorders and cancers. Given TBCK’s diverse roles, we have focused on its putative kinase domain. Through comprehensive analysis using tools such as Kinase Tree, AlphaFold2, Clustal Omega, and SMART, we discovered that TBCK lacks the key “D” residue in the conserved “HRD” and “DFG” motifs typical of protein kinases like PKA and SRC, suggesting TBCK functions as a pseudokinase. Intriguingly, gene ontology analysis from recent RNA-seq data indicates TBCK’s involvement in regulating protein phosphorylation. This suggests that TBCK may influence protein phosphorylation either directly through its potential kinase domain or indirectly via interactions with other proteins. To uncover the full spectrum of TBCK’s roles in neurogenetic disorders and cancer, urgent high-throughput analyses are necessary to identify its interacting partners.