要約
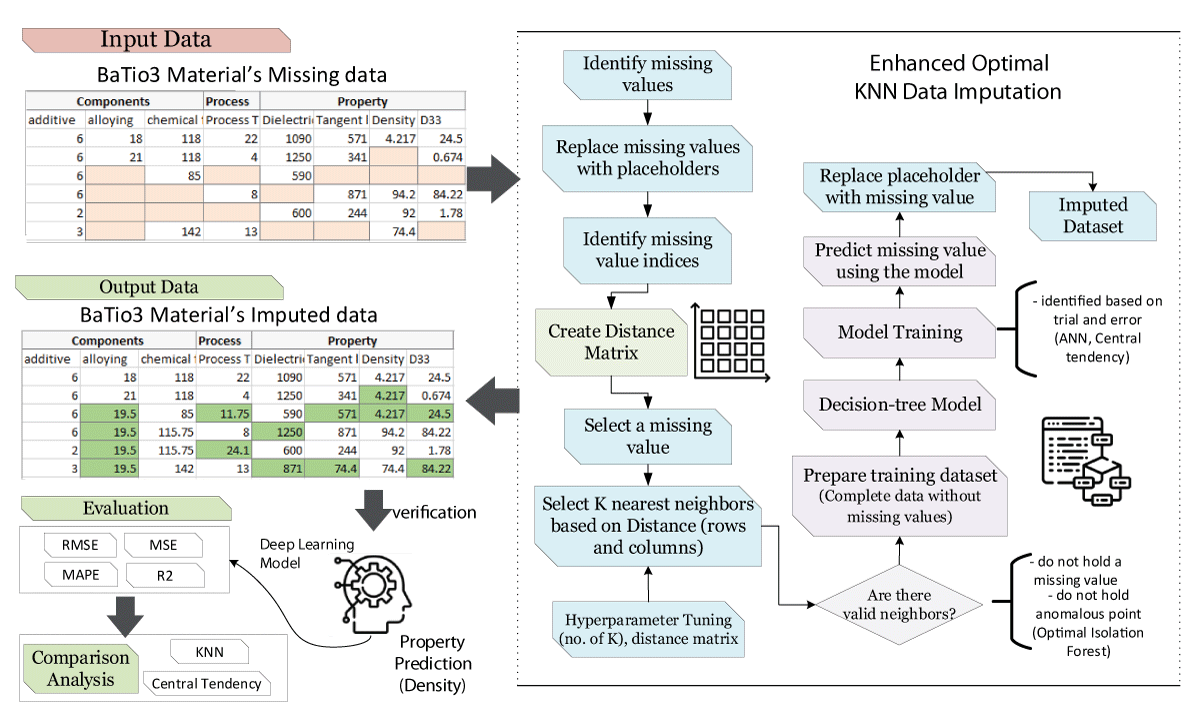
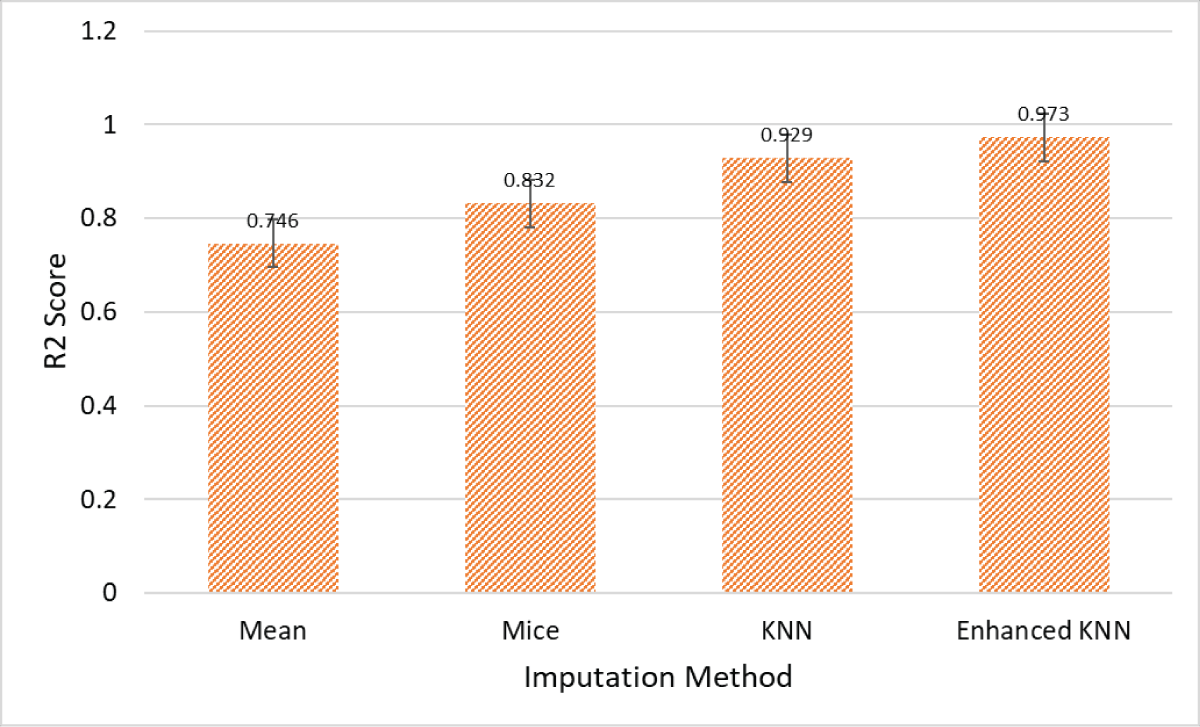
In materials science, the integrity and completeness of datasets are critical for robust predictive modeling. Unfortunately, material datasets frequently contain missing values due to factors such as measurement errors, data non-availability, or experimental limitations, which can significantly undermine the accuracy of property predictions. To tackle this challenge, we introduce an optimized K-Nearest Neighbors (KNN) imputation method, augmented with Deep Neural Network (DNN) modeling, to enhance the accuracy of predicting material properties. Our study compares the performance of our Enhanced KNN method against traditional imputation techniques—mean imputation and Multiple Imputation by Chained Equations (MICE). The results indicate that our Enhanced KNN method achieves a superior R² score of 0.973, which represents a significant improvement of 0.227 over Mean imputation, 0.141 over MICE, and 0.044 over KNN imputation. This enhancement not only boosts the data integrity but also preserves the statistical characteristics essential for reliable predictions in materials science.